Hardware requirements and performance benchmark
Summary
For the preprocessing step, the rnaends package makes use of fastqStreamer from the ShortRead package. Thus, the yieldSize parameter allows to reduce memory requirement as only a certain number of reads will be processed at a time (default is set to 1 million in rnaends).
For the mapping of RNA-Seq reads to the reference sequences, the rnaends package simply uses on the wrappers Rbowtie and Rsubread, respectively for the bowtie and subread aligners.
For quantification step, the count table (internally stored as a tibble), and the downstream analyses, the memory needed depends mainly on the genome or reference sequences studied (i.e. the number of positions corresponding to 5’ and/or 3’ ends observed in the RNA-Seq reads). Thus, the memory needed scales linearly with the length of the reference sequences and the number of experiments/samples/conditions.
Note
Conclusion: All the notebooks presented in the vignettes, and this benchmark can run on a laptop with 16GB memory.
The running times and memory requirements scale linearly
with the number of sequencing reads for the pre-processing steps and their mapping to references sequences,
with the number of 5’/3’ end locations and samples in the count table.
Material and methods
Hardware: The following was performed on a Dell Latitude 5280 laptop with dual core CPU Intel i5-7300 with 16GB memory.
The package peakRAM was chosen as it intends to capture real memory requirements within R, whereas others generally only capture memory allocations from R functions and data structures (but not compiled code). From the authors: «So-called “copy-on-modify” behavior, characteristic of R, means that some expressions or functions may require an unexpectedly large amount of RAM overhead. For example, replacing a single value in a matrix (e.g., with ‘[<-’) duplicates that matrix in the backend, making this task require twice as much RAM as that used by the matrix itself.»
library(pryr)
library(peakRAM)
library(tidyverse)
library(ggpubr)
mem_used()
#> 90.9 MB
library(rnaends)
mem_used()
#> 813 MB
Dataset
FASTQ files with variable sequencing depths (1 to 20 millions of reads per experimental condition) were generated based on observed distributions of 5’ ends in two conditions (5’PPP only RNAs vs. background noise as in the TSS-EMOTE protocol) to evaluate the performances of different features of the rnaends package:
pre-processing of raw reads
only validate then extract the mappable part of reads.
checking the presence and validity of some sequences, i.e. a recognition sequence (CGGCACCAACCGAGG) and a control sequence (CGC) at precise locations on the reads (see the read structure below for details).
checking the validity of UMIs and extracting them for further use for PCR duplicates removal.
using barcodes for demultiplexing experimental conditions.
quantification
mapping
count table
downstream
TSS
100 nucleotide long reads were generated.
UMIs are 17 nucleotides long and are composed of only A, C and G.
12 barcodes were used corresponding to two experimental conditions with 3 or 6 replicates depending on the fact that the first 2 or 3 nucleotides of the barcodes are used:
AAxx, ACxx, ATxx for 3 replicates of the 5’PPP RNAs condition, and AAAx, AACx, ACAx, ACCx, ATAx, ATCx for the 6 replicates
CAxx, CCxx, CTxx for the 3 replicates of the background noise condition, and CAAx, CACx, CCAx, CCCx, CTAx, CTCx for the 6 replicates.
As a result, benchmarks for 12 barcodes process 12 times 1-5-10-15-20 million reads per barcode/condition, and benchmarks with 6 barcodes process 2-10-20-30-40 million reads per barcode/condition. Thus, the biggest analysis presented here (6 barcodes with 20 millions reads per condition), consists in 240 million reads.
Dataset availability: FASTQ files used in this benchmark are available on zenodo at https://doi.org/10.5281/zenodo.17592454
Pre-processing of reads
Reads Parsing and Validation: parse_reads
The reads in the raw FASTQ files have the following read structure:
1 2 5 6 20 21 37 38 40 41 100
N - barcode - CGGCACCAACCGAGG - VVVVVVV - CGC - XXXXXXXXXXXXXXXXXXXXX
1 random nt - barcode - recognition.seq - UMI - control.seq - RNA 5' end
FASTQ files with 1 to 20 millions reads:
fq_file_list <- c("prepro.UMI-RC-CS.1M.fq.gz",
"prepro.UMI-RC-CS.5M.fq.gz",
"prepro.UMI-RC-CS.10M.fq.gz",
"prepro.UMI-RC-CS.15M.fq.gz",
"prepro.UMI-RC-CS.20M.fq.gz"
)
Read features only with a mappable part to validate and extract: parse_reads
Read features with only the mappable part to extract
rfwo <- init_read_features(start=41, width=60)
rfwo %>%
knitr::kable()
name |
start |
width |
pattern_type |
pattern |
max_mismatch |
readid_prepend |
|---|---|---|---|---|---|---|
readseq |
41 |
60 |
nucleotides |
ACTG |
0 |
FALSE |
Measure time and maximum RAM in use for the parse_read function:
prwo <- sapply(fq_file_list, function(i) peakRAM(
parse_reads(fastq_file = i,
features = rfwo,
force = TRUE)
)
)
prwo <- prwo %>%
t %>%
as_tibble() %>%
select(time=Elapsed_Time_sec,
peak_ram=Peak_RAM_Used_MiB) %>%
mutate(time=duration(as.numeric(time), units="seconds"),
peak_ram = as.numeric(peak_ram),
nb_reads = c(1, 5, 10, 15, 20),
features = "none")
prwo %>%
knitr::kable()
time |
peak_ram |
nb_reads |
features |
|---|---|---|---|
27.355s |
470 |
1 |
none |
127.94s (~2.13 minutes) |
907 |
5 |
none |
253.563s (~4.23 minutes) |
1010 |
10 |
none |
378.18s (~6.3 minutes) |
1086 |
15 |
none |
505.403s (~8.42 minutes) |
1202 |
20 |
none |
Read features with a recognition sequence and a control sequence: parse_reads
Read features with a recognition sequence and a control sequence:
rfw_rs <- add_read_feature(rfwo,
name="recog",
start=6,
width=15,
pattern = "CGGCACCAACCGAGG",
pattern_type = "constant")
rfw_rs <- add_read_feature(rfw_rs,
name="ctrl",
start=38,
width=3,
pattern = "CGC",
pattern_type = "constant")
rfw_rs %>%
knitr::kable()
name |
start |
width |
pattern_type |
pattern |
max_mismatch |
readid_prepend |
|---|---|---|---|---|---|---|
readseq |
41 |
60 |
nucleotides |
ACTG |
0 |
FALSE |
recog |
6 |
15 |
constant |
CGGCACCA…. |
0 |
FALSE |
ctrl |
38 |
3 |
constant |
CGC |
0 |
FALSE |
Measure time and maximum RAM in use for the parse_read function:
prw_rs <- sapply(fq_file_list, function(i) peakRAM(
parse_reads(fastq_file = i,
features = rfw_rs,
force = TRUE)
)
)
prw_rs <- prw_rs %>%
t %>%
as_tibble() %>%
select(time=Elapsed_Time_sec,
peak_ram=Peak_RAM_Used_MiB) %>%
mutate(time=duration(as.numeric(time), units="seconds"),
peak_ram = as.numeric(peak_ram),
nb_reads = c(1, 5, 10, 15, 20),
features = "recognition and control seq")
prw_rs %>%
knitr::kable()
time |
peak_ram |
nb_reads |
features |
|---|---|---|---|
25.4450000000002s |
526 |
1 |
recognition and control seq |
128.704s (~2.15 minutes) |
1247 |
5 |
recognition and control seq |
255.521s (~4.26 minutes) |
1086 |
10 |
recognition and control seq |
384.678s (~6.41 minutes) |
1166 |
15 |
recognition and control seq |
545.71s (~9.1 minutes) |
1046 |
20 |
recognition and control seq |
Read features with UMIs: parse_reads
Read features with UMIs:
rfw_umi <- add_read_feature(rfwo,
name="UMI",
start=21,
width=17,
pattern="ACG",
pattern_type="nucleotides",
readid_prepend = TRUE)
rfw_umi %>%
knitr::kable()
name |
start |
width |
pattern_type |
pattern |
max_mismatch |
readid_prepend |
|---|---|---|---|---|---|---|
readseq |
41 |
60 |
nucleotides |
ACTG |
0 |
FALSE |
UMI |
21 |
17 |
nucleotides |
ACG |
0 |
TRUE |
Measure time and maximum RAM in use for the parse_read function:
prw_umi <- sapply(fq_file_list, function(i) peakRAM(
parse_reads(fastq_file = i,
features = rfw_umi,
force = TRUE)
)
)
prw_umi <- prw_umi %>%
t %>%
as_tibble() %>%
select(time=Elapsed_Time_sec,
peak_ram=Peak_RAM_Used_MiB) %>%
mutate(time=duration(as.numeric(time), units="seconds"),
peak_ram = as.numeric(peak_ram),
nb_reads = c(1, 5, 10, 15, 20),
features = "UMI")
prw_umi %>%
knitr::kable()
time |
peak_ram |
nb_reads |
features |
|---|---|---|---|
33.8269999999998s |
636 |
1 |
UMI |
167.811s (~2.8 minutes) |
1030 |
5 |
UMI |
343.696s (~5.73 minutes) |
1110 |
10 |
UMI |
514.835s (~8.58 minutes) |
1192 |
15 |
UMI |
669.22s (~11.15 minutes) |
1280 |
20 |
UMI |
Summary
pr <- bind_rows(prwo, prw_rs, prw_umi)
Running time and memory footprint depending on the number of reads:
p1 <- pr %>%
ggplot() +
geom_line(aes(x=nb_reads, y=time, color=features),
show.legend = FALSE) +
geom_point(aes(x=nb_reads, y=time, shape=features, color=features),
show.legend = FALSE) +
theme_light() +
xlab("Number of reads (in millions)") +
ylab("Duration (in seconds)") +
ggtitle("Running times for pre-processing functions")
p2 <- pr %>%
ggplot() +
geom_line(aes(x=nb_reads, y=peak_ram, color=features)) +
geom_point(aes(x=nb_reads, y=peak_ram, shape=features, color=features)) +
theme_light() +
xlab("Number of reads (in millions)") +
ylab("Memory used (in MB)") +
ggtitle("Memory used during pre-processing functions") +
theme(legend.position="inside", legend.position.inside = c(0.6, 0.15))
ggarrange(p1, p2)
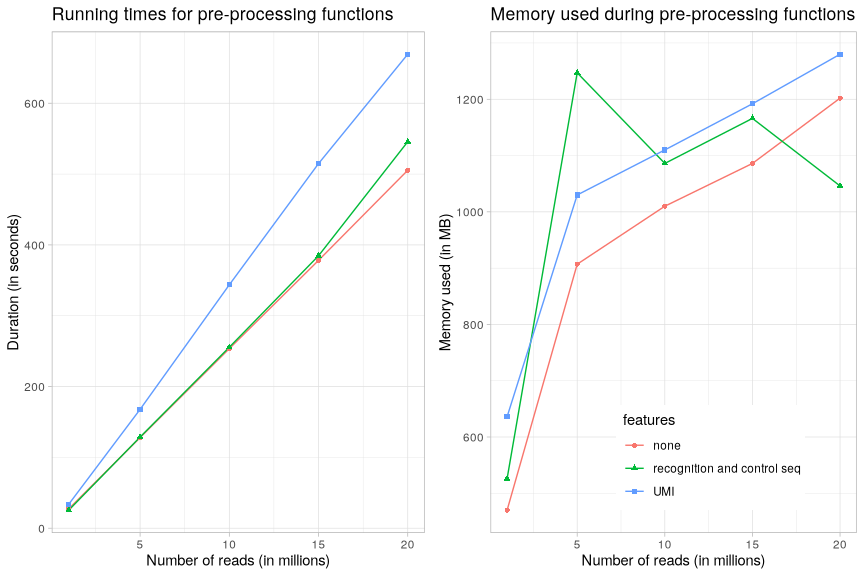
Note
In the above graphs, we can see that the running time scales linearly with the number of sequencing reads to analyse. The overhead for the validation of constant strings at certain locations in reads (i.e. recognition and control sequences) is negligible. The UMI validation and extraction for further analyses scale also linearly with a substantial overhead.
Regarding memory footprint, we can observe that, except for small amounts of reads (below 5 millions), the memory in use is aroung 1-1.5GB. This is to be correlated with the yieldSize parameter (default: 1 million) used by the fastqStreamer from the ShortRead package for processing reads, i.e. reads are loaded and processed by groups of 1 million reads from the FASTQ file, thus only a certain amount of memory is necessary.
Reads Parsing and demultiplexing: demultiplex
Read features with barcodes for demultiplexing: demultiplex
FASTQ files composed of multiplexed FASTQ files with various sequencing depths. 1M means 1 million reads per condition, thus 12 millions reads in total:
fq_files_bc <- c("prepro.PPP-BG.1M.fq.gz",
"prepro.PPP-BG.5M.fq.gz",
"prepro.PPP-BG.10M.fq.gz",
"prepro.PPP-BG.15M.fq.gz",
"prepro.PPP-BG.20M.fq.gz"
)
Read features with barcodes of length 2 and 6 experiments: 3 replicates for control samples (background noise), and 3 replicates for test samples (5’PPP samples)
rfw_bc2 <- add_read_feature(rfwo,
name="barcode",
start=2,
width=2,
pattern=c("AA","AC","AT","CA","CC","CT"),
pattern_type = "constant",
readid_prepend = FALSE
)
rfw_bc2 %>%
knitr::kable()
name |
start |
width |
pattern_type |
pattern |
max_mismatch |
readid_prepend |
|---|---|---|---|---|---|---|
readseq |
41 |
60 |
nucleotides |
ACTG |
0 |
FALSE |
barcode |
2 |
2 |
constant |
AA, AC, …. |
0 |
FALSE |
Measure time and maximum RAM in use for the demultiplex function for 6 barcodes:
prw_bc2 <- sapply(fq_files_bc, function(i) peakRAM(
demultiplex(fastq_file = i,
features = rfw_bc2,
force = TRUE)
)
)
prw_bc2 <- prw_bc2 %>%
t %>%
as_tibble() %>%
select(time=Elapsed_Time_sec,
peak_ram=Peak_RAM_Used_MiB) %>%
mutate(time=duration(as.numeric(time), units="seconds"),
peak_ram = as.numeric(peak_ram),
nb_reads = c(12, 60, 120, 180, 240),
features = "6 barcodes")
prw_bc2 %>%
knitr::kable()
time |
peak_ram |
nb_reads |
features |
|---|---|---|---|
321.641000000001s (~5.36 minutes) |
1098 |
12 |
6 barcodes |
1602.599s (~26.71 minutes) |
1182 |
60 |
6 barcodes |
4291.41s (~1.19 hours) |
1035 |
120 |
6 barcodes |
4720.811s (~1.31 hours) |
1004 |
180 |
6 barcodes |
6297.646s (~1.75 hours) |
1004 |
240 |
6 barcodes |
Read features with barcodes of length 3 and 12 experiments: 6 replicates for control samples (background noise), and 6 replicates for test samples (5’PPP samples)
rfw_bc3 <- add_read_feature(rfwo,
name="barcode",
start=2,
width=3,
pattern=c("AAA","AAC","ACA","ACC","ATA","ATC",
"CAA","CAC","CCA","CCC","CTA","CTC"),
pattern_type = "constant",
readid_prepend = FALSE
)
rfw_bc3 %>%
knitr::kable()
name |
start |
width |
pattern_type |
pattern |
max_mismatch |
readid_prepend |
|---|---|---|---|---|---|---|
readseq |
41 |
60 |
nucleotides |
ACTG |
0 |
FALSE |
barcode |
2 |
3 |
constant |
AAA, AAC…. |
0 |
FALSE |
Measure time and maximum RAM in use for the demultiplex function for 12 barcodes:
prw_bc3 <- sapply(fq_files_bc, function(i) peakRAM(
demultiplex(fastq_file = i,
features = rfw_bc3,
force = TRUE)
)
)
prw_bc3 <- prw_bc3 %>%
t %>%
as_tibble() %>%
select(time=Elapsed_Time_sec,
peak_ram=Peak_RAM_Used_MiB) %>%
mutate(time=duration(as.numeric(time), units="seconds"),
peak_ram = as.numeric(peak_ram),
nb_reads = c(12, 60, 120, 180, 240),
features = "12 barcodes")
prw_bc3 %>%
knitr::kable()
time |
peak_ram |
nb_reads |
features |
|---|---|---|---|
316.102999999999s (~5.27 minutes) |
1005 |
12 |
12 barcodes |
1589.093s (~26.48 minutes) |
1005 |
60 |
12 barcodes |
3158.782s (~52.65 minutes) |
1247 |
120 |
12 barcodes |
4731.018s (~1.31 hours) |
1084 |
180 |
12 barcodes |
6256.014s (~1.74 hours) |
1165 |
240 |
12 barcodes |
Read features combined: demultiplex
Read features with recognition and control sequences, UMIs, and barcodes:
rfw_a <- add_read_feature(rfwo,
name="recog",
start=6,
width=15,
pattern = "CGGCACCAACCGAGG",
pattern_type = "constant")
rfw_a <- add_read_feature(rfw_a,
name="ctrl",
start=38,
width=3,
pattern = "CGC",
pattern_type = "constant")
rfw_a <- add_read_feature(rfw_a,
name="UMI",
start=21,
width=17,
pattern="ACG",
pattern_type="nucleotides",
readid_prepend = TRUE)
rfw_a <- add_read_feature(rfw_a,
name="barcode",
start=2,
width=2,
pattern=c("AA","AC","AT","CA","CC","CT"),
pattern_type = "constant",
readid_prepend = FALSE)
rfw_a %>%
knitr::kable()
name |
start |
width |
pattern_type |
pattern |
max_mismatch |
readid_prepend |
|---|---|---|---|---|---|---|
readseq |
41 |
60 |
nucleotides |
ACTG |
0 |
FALSE |
recog |
6 |
15 |
constant |
CGGCACCA…. |
0 |
FALSE |
ctrl |
38 |
3 |
constant |
CGC |
0 |
FALSE |
UMI |
21 |
17 |
nucleotides |
ACG |
0 |
TRUE |
barcode |
2 |
2 |
constant |
AA, AC, …. |
0 |
FALSE |
Measure time and maximum RAM in use for the demultiplex function for combined read features:
prw_a <- sapply(fq_files_bc, function(i) peakRAM(
demultiplex(fastq_file = i,
features = rfw_a,
force = TRUE)
)
)
prw_a <- prw_a %>%
t %>%
as_tibble() %>%
select(time=Elapsed_Time_sec,
peak_ram=Peak_RAM_Used_MiB) %>%
mutate(time=duration(as.numeric(time), units="seconds"),
peak_ram = as.numeric(peak_ram),
nb_reads = c(12, 60, 120, 180, 240),
features = "RS+CS, UMI, 6 barcodes")
prw_a %>%
knitr::kable()
time |
peak_ram |
nb_reads |
features |
|---|---|---|---|
401.449999999997s (~6.69 minutes) |
1252 |
12 |
RS+CS, UMI, 6 barcodes |
1992.30899999999s (~33.21 minutes) |
1344 |
60 |
RS+CS, UMI, 6 barcodes |
3932.47199999999s (~1.09 hours) |
1448 |
120 |
RS+CS, UMI, 6 barcodes |
5881.689s (~1.63 hours) |
1238 |
180 |
RS+CS, UMI, 6 barcodes |
7933s (~2.2 hours) |
1355 |
240 |
RS+CS, UMI, 6 barcodes |
Summary
pr2 <- bind_rows(prw_bc2, prw_bc3, prw_a)
Running time and memory footprint depending on the number of reads:
p1 <- pr2 %>%
ggplot() +
geom_line(aes(x=nb_reads, y=time, color=features)) +
geom_point(aes(x=nb_reads, y=time, shape=features, color=features)) +
theme_light() +
xlab("Number of reads (in millions)") +
ylab("Duration (in seconds)") +
ggtitle("Running times for demultipexing") +
theme(legend.position="inside", legend.position.inside = c(0.6, 0.15))
p2 <- pr2 %>%
ggplot() +
geom_line(aes(x=nb_reads, y=peak_ram, color=features),
show.legend = FALSE) +
geom_point(aes(x=nb_reads, y=peak_ram, shape=features, color=features),
show.legend = FALSE) +
theme_light() +
xlab("Number of reads (in millions)") +
ylab("Memory used (in MB)") +
ggtitle("Memory used during demultipexing")
ggarrange(p1, p2)
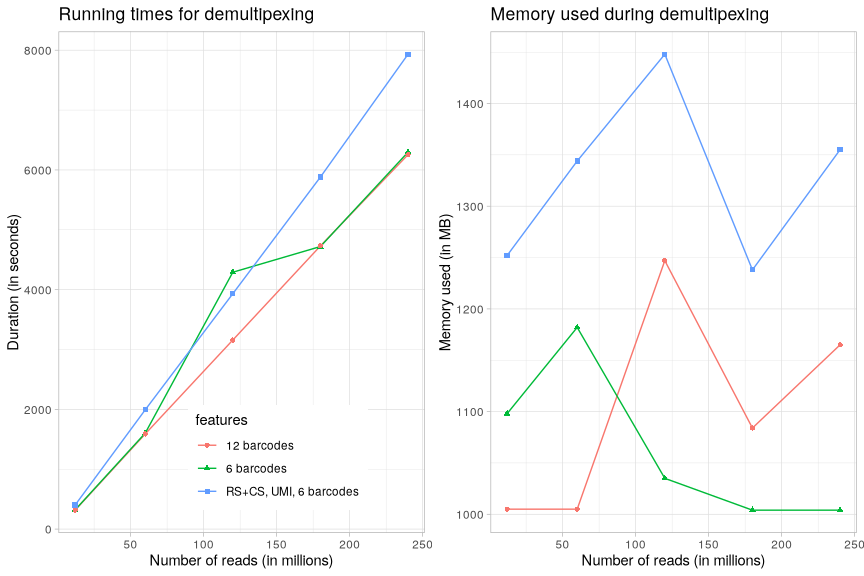
Note
In the above graphs, we can see that the running times for demultiplexing scale linearly with the number of reads to analyse, regardless of the number of analyses to demultiplex. As in the previous section, the overhead of additional features to parse, validate and extract (recogntion sequence denoted RS, control sequence denoted CS, and UMIs) scale also linaerly with the size of the input.
Regarding the memory footprint, we observe the same behavior as in the previous section, i.e. it mainly depends on the yieldSize parameter used for processing reads per chunk by the fastqStreamer from the ShortRead package.
Quantification
Data preparation: use previous read features with combined features.
dirout <- c("quantif.1M", "quantif.5M", "quantif.10M", "quantif.15M", "quantif.20M")
names(dirout) <- fq_files_bc
sapply(fq_files_bc, function(i)
demultiplex(fastq_file = i,
features = rfw_a,
out_dir = dirout[i],
force = FALSE)
)
#> prepro.PPP-BG.1M.fq.gz prepro.PPP-BG.5M.fq.gz
#> barcode_group character,6 character,6
#> is_valid_readseq logical,6 logical,6
#> is_valid_recog logical,6 logical,6
#> is_valid_ctrl logical,6 logical,6
#> is_valid_UMI logical,6 logical,6
#> is_valid_barcode logical,6 logical,6
#> count integer,6 integer,6
#> demux_filename character,6 character,6
#> prepro.PPP-BG.10M.fq.gz prepro.PPP-BG.15M.fq.gz
#> barcode_group character,6 character,6
#> is_valid_readseq logical,6 logical,6
#> is_valid_recog logical,6 logical,6
#> is_valid_ctrl logical,6 logical,6
#> is_valid_UMI logical,6 logical,6
#> is_valid_barcode logical,6 logical,6
#> count integer,6 integer,6
#> demux_filename character,6 character,6
#> prepro.PPP-BG.20M.fq.gz
#> barcode_group character,6
#> is_valid_readseq logical,6
#> is_valid_recog logical,6
#> is_valid_ctrl logical,6
#> is_valid_UMI logical,6
#> is_valid_barcode logical,6
#> count integer,6
#> demux_filename character,6
Mapping: map_fastq_files
Measure time and maximum RAM in use for the map_fastq_files function with bowtie:
prw_bt <- sapply(dirout, function(i) peakRAM(
map_fastq_files(
list_fastq_files = list.files(i,
pattern = "_valid.fastq.gz$",
full.names = TRUE),
index_name = "MW2.bt",
fasta_name = "MW2.fasta",
max_map_mismatch = 1,
aligner = "bowtie",
map_unique = TRUE,
force = TRUE
))
)
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
#> Warning in FUN(X[[i]], ...): Index directory already exists remove it if you
#> want to recreate it
prw_bt <- prw_bt %>%
t %>%
as_tibble() %>%
select(time=Elapsed_Time_sec,
peak_ram=Peak_RAM_Used_MiB) %>%
mutate(time=duration(as.numeric(time), units="seconds"),
peak_ram = as.numeric(peak_ram),
nb_reads = c(12, 60, 120, 180, 240),
aligner = "bowtie")
prw_bt %>%
knitr::kable()
time |
peak_ram |
nb_reads |
features |
|---|---|---|---|
229.714000000007s (~3.83 minutes) |
1.2 |
12 |
bowtie |
1192.98299999999s (~19.88 minutes) |
0.5 |
60 |
bowtie |
2357.016s (~39.28 minutes) |
0.5 |
120 |
bowtie |
3545.39999999999s (~59.09 minutes) |
0.5 |
180 |
bowtie |
4709.34299999999s (~1.31 hours) |
0.5 |
240 |
bowtie |
Measure time and maximum RAM in use for the map_fastq_files function with subread:
prw_sr <- sapply(dirout, function(i) peakRAM(
map_fastq_files(
list_fastq_files = list.files(i,
pattern = "_valid.fastq.gz$",
full.names = TRUE),
index_name = "MW2.sr",
fasta_name = "MW2.fasta",
max_map_mismatch = 1,
aligner = "subread",
map_unique = TRUE,
force = TRUE
))
)
subread output
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.1M_AA_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.1M_AA_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:32:38, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=73.5k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=102.7k reads per second ||
#> || 13% completed, 1.2 mins elapsed, rate=104.6k reads per second ||
#> || 19% completed, 1.2 mins elapsed, rate=105.4k reads per second ||
#> || 26% completed, 1.2 mins elapsed, rate=105.0k reads per second ||
#> || 33% completed, 1.3 mins elapsed, rate=103.9k reads per second ||
#> || 39% completed, 1.3 mins elapsed, rate=104.4k reads per second ||
#> || 46% completed, 1.3 mins elapsed, rate=104.5k reads per second ||
#> || 53% completed, 1.3 mins elapsed, rate=103.4k reads per second ||
#> || 59% completed, 1.3 mins elapsed, rate=103.2k reads per second ||
#> || 66% completed, 1.4 mins elapsed, rate=103.3k reads per second ||
#> || 70% completed, 1.4 mins elapsed, rate=16.9k reads per second ||
#> || 73% completed, 1.4 mins elapsed, rate=17.5k reads per second ||
#> || 76% completed, 1.4 mins elapsed, rate=18.1k reads per second ||
#> || 79% completed, 1.4 mins elapsed, rate=18.7k reads per second ||
#> || 83% completed, 1.4 mins elapsed, rate=19.2k reads per second ||
#> || 86% completed, 1.5 mins elapsed, rate=19.8k reads per second ||
#> || 90% completed, 1.5 mins elapsed, rate=20.4k reads per second ||
#> || 93% completed, 1.5 mins elapsed, rate=20.9k reads per second ||
#> || 96% completed, 1.5 mins elapsed, rate=21.4k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 2,000,000 ||
#> || Mapped : 1,422,829 (71.1%) ||
#> || Uniquely mapped : 1,422,829 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 577,171 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 1.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.1M_AC_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.1M_AC_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:34:13, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=102.4k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=109.0k reads per second ||
#> || 13% completed, 1.2 mins elapsed, rate=109.0k reads per second ||
#> || 19% completed, 1.2 mins elapsed, rate=108.8k reads per second ||
#> || 26% completed, 1.2 mins elapsed, rate=108.9k reads per second ||
#> || 33% completed, 1.2 mins elapsed, rate=108.9k reads per second ||
#> || 39% completed, 1.3 mins elapsed, rate=109.1k reads per second ||
#> || 46% completed, 1.3 mins elapsed, rate=109.0k reads per second ||
#> || 53% completed, 1.3 mins elapsed, rate=108.8k reads per second ||
#> || 59% completed, 1.3 mins elapsed, rate=108.7k reads per second ||
#> || 66% completed, 1.3 mins elapsed, rate=108.5k reads per second ||
#> || 69% completed, 1.4 mins elapsed, rate=17.1k reads per second ||
#> || 73% completed, 1.4 mins elapsed, rate=17.7k reads per second ||
#> || 76% completed, 1.4 mins elapsed, rate=18.3k reads per second ||
#> || 79% completed, 1.4 mins elapsed, rate=18.9k reads per second ||
#> || 83% completed, 1.4 mins elapsed, rate=19.4k reads per second ||
#> || 86% completed, 1.4 mins elapsed, rate=20.0k reads per second ||
#> || 89% completed, 1.5 mins elapsed, rate=20.6k reads per second ||
#> || 93% completed, 1.5 mins elapsed, rate=21.1k reads per second ||
#> || 96% completed, 1.5 mins elapsed, rate=21.7k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 2,000,000 ||
#> || Mapped : 1,423,160 (71.2%) ||
#> || Uniquely mapped : 1,423,160 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 576,840 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 1.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.1M_AT_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.1M_AT_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:35:48, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=99.3k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=109.3k reads per second ||
#> || 13% completed, 1.2 mins elapsed, rate=107.0k reads per second ||
#> || 19% completed, 1.2 mins elapsed, rate=107.9k reads per second ||
#> || 26% completed, 1.2 mins elapsed, rate=108.6k reads per second ||
#> || 32% completed, 1.3 mins elapsed, rate=108.8k reads per second ||
#> || 39% completed, 1.3 mins elapsed, rate=108.9k reads per second ||
#> || 46% completed, 1.3 mins elapsed, rate=109.0k reads per second ||
#> || 52% completed, 1.3 mins elapsed, rate=109.0k reads per second ||
#> || 59% completed, 1.3 mins elapsed, rate=109.1k reads per second ||
#> || 66% completed, 1.4 mins elapsed, rate=108.6k reads per second ||
#> || 70% completed, 1.4 mins elapsed, rate=16.9k reads per second ||
#> || 73% completed, 1.4 mins elapsed, rate=17.5k reads per second ||
#> || 76% completed, 1.4 mins elapsed, rate=18.2k reads per second ||
#> || 79% completed, 1.4 mins elapsed, rate=18.7k reads per second ||
#> || 83% completed, 1.4 mins elapsed, rate=19.3k reads per second ||
#> || 86% completed, 1.5 mins elapsed, rate=19.9k reads per second ||
#> || 89% completed, 1.5 mins elapsed, rate=20.4k reads per second ||
#> || 93% completed, 1.5 mins elapsed, rate=21.0k reads per second ||
#> || 96% completed, 1.5 mins elapsed, rate=21.5k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 2,000,000 ||
#> || Mapped : 1,424,301 (71.2%) ||
#> || Uniquely mapped : 1,424,301 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 575,699 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 1.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.1M_CA_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.1M_CA_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:37:23, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=105.9k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=114.8k reads per second ||
#> || 13% completed, 1.2 mins elapsed, rate=115.5k reads per second ||
#> || 20% completed, 1.2 mins elapsed, rate=115.4k reads per second ||
#> || 26% completed, 1.2 mins elapsed, rate=115.2k reads per second ||
#> || 33% completed, 1.2 mins elapsed, rate=115.0k reads per second ||
#> || 40% completed, 1.3 mins elapsed, rate=115.0k reads per second ||
#> || 46% completed, 1.3 mins elapsed, rate=115.1k reads per second ||
#> || 53% completed, 1.3 mins elapsed, rate=115.1k reads per second ||
#> || 60% completed, 1.3 mins elapsed, rate=114.8k reads per second ||
#> || 70% completed, 1.4 mins elapsed, rate=17.1k reads per second ||
#> || 73% completed, 1.4 mins elapsed, rate=17.7k reads per second ||
#> || 76% completed, 1.4 mins elapsed, rate=18.3k reads per second ||
#> || 80% completed, 1.4 mins elapsed, rate=18.8k reads per second ||
#> || 83% completed, 1.4 mins elapsed, rate=19.4k reads per second ||
#> || 86% completed, 1.5 mins elapsed, rate=19.9k reads per second ||
#> || 89% completed, 1.5 mins elapsed, rate=20.4k reads per second ||
#> || 93% completed, 1.5 mins elapsed, rate=20.9k reads per second ||
#> || 96% completed, 1.5 mins elapsed, rate=21.4k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 2,000,000 ||
#> || Mapped : 374,571 (18.7%) ||
#> || Uniquely mapped : 374,571 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 1,625,429 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 1.7 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.1M_CC_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.1M_CC_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:39:02, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=102.4k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=113.6k reads per second ||
#> || 13% completed, 1.2 mins elapsed, rate=114.8k reads per second ||
#> || 20% completed, 1.2 mins elapsed, rate=115.3k reads per second ||
#> || 26% completed, 1.2 mins elapsed, rate=115.7k reads per second ||
#> || 33% completed, 1.2 mins elapsed, rate=115.7k reads per second ||
#> || 40% completed, 1.3 mins elapsed, rate=115.9k reads per second ||
#> || 46% completed, 1.3 mins elapsed, rate=116.0k reads per second ||
#> || 53% completed, 1.3 mins elapsed, rate=116.1k reads per second ||
#> || 60% completed, 1.3 mins elapsed, rate=115.5k reads per second ||
#> || 66% completed, 1.3 mins elapsed, rate=115.6k reads per second ||
#> || 70% completed, 1.4 mins elapsed, rate=17.1k reads per second ||
#> || 73% completed, 1.4 mins elapsed, rate=17.7k reads per second ||
#> || 76% completed, 1.4 mins elapsed, rate=18.3k reads per second ||
#> || 80% completed, 1.4 mins elapsed, rate=18.8k reads per second ||
#> || 83% completed, 1.4 mins elapsed, rate=19.4k reads per second ||
#> || 86% completed, 1.5 mins elapsed, rate=19.9k reads per second ||
#> || 90% completed, 1.5 mins elapsed, rate=20.4k reads per second ||
#> || 93% completed, 1.5 mins elapsed, rate=20.9k reads per second ||
#> || 96% completed, 1.5 mins elapsed, rate=21.4k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 2,000,000 ||
#> || Mapped : 375,407 (18.8%) ||
#> || Uniquely mapped : 375,407 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 1,624,593 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 1.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.1M_CT_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.1M_CT_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:40:37, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=103.0k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=115.3k reads per second ||
#> || 13% completed, 1.2 mins elapsed, rate=115.6k reads per second ||
#> || 20% completed, 1.2 mins elapsed, rate=115.9k reads per second ||
#> || 26% completed, 1.2 mins elapsed, rate=116.0k reads per second ||
#> || 33% completed, 1.2 mins elapsed, rate=115.9k reads per second ||
#> || 40% completed, 1.3 mins elapsed, rate=116.1k reads per second ||
#> || 46% completed, 1.3 mins elapsed, rate=116.0k reads per second ||
#> || 53% completed, 1.3 mins elapsed, rate=115.5k reads per second ||
#> || 60% completed, 1.3 mins elapsed, rate=115.5k reads per second ||
#> || 69% completed, 1.4 mins elapsed, rate=17.1k reads per second ||
#> || 73% completed, 1.4 mins elapsed, rate=17.7k reads per second ||
#> || 76% completed, 1.4 mins elapsed, rate=18.3k reads per second ||
#> || 80% completed, 1.4 mins elapsed, rate=18.8k reads per second ||
#> || 83% completed, 1.4 mins elapsed, rate=19.4k reads per second ||
#> || 86% completed, 1.5 mins elapsed, rate=19.9k reads per second ||
#> || 90% completed, 1.5 mins elapsed, rate=20.4k reads per second ||
#> || 93% completed, 1.5 mins elapsed, rate=20.9k reads per second ||
#> || 96% completed, 1.5 mins elapsed, rate=21.4k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 2,000,000 ||
#> || Mapped : 374,947 (18.7%) ||
#> || Uniquely mapped : 374,947 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 1,625,053 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 1.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.5M_AA_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.5M_AA_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:42:14, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=98.2k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=107.9k reads per second ||
#> || 12% completed, 1.3 mins elapsed, rate=107.5k reads per second ||
#> || 19% completed, 1.4 mins elapsed, rate=107.3k reads per second ||
#> || 26% completed, 1.6 mins elapsed, rate=107.2k reads per second ||
#> || 32% completed, 1.7 mins elapsed, rate=106.7k reads per second ||
#> || 39% completed, 1.8 mins elapsed, rate=105.9k reads per second ||
#> || 46% completed, 1.9 mins elapsed, rate=105.2k reads per second ||
#> || 48% completed, 1.9 mins elapsed, rate=41.9k reads per second ||
#> || 51% completed, 2.0 mins elapsed, rate=42.6k reads per second ||
#> || 53% completed, 2.1 mins elapsed, rate=43.3k reads per second ||
#> || 55% completed, 2.1 mins elapsed, rate=44.1k reads per second ||
#> || 58% completed, 2.2 mins elapsed, rate=44.7k reads per second ||
#> || 60% completed, 2.2 mins elapsed, rate=45.4k reads per second ||
#> || 62% completed, 2.3 mins elapsed, rate=45.9k reads per second ||
#> || 65% completed, 2.3 mins elapsed, rate=46.4k reads per second ||
#> || 67% completed, 2.4 mins elapsed, rate=46.8k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 3.2 mins elapsed, rate=56.9k reads per second ||
#> || 76% completed, 3.4 mins elapsed, rate=55.6k reads per second ||
#> || 83% completed, 3.6 mins elapsed, rate=57.1k reads per second ||
#> || 90% completed, 3.7 mins elapsed, rate=40.8k reads per second ||
#> || 91% completed, 3.7 mins elapsed, rate=41.0k reads per second ||
#> || 92% completed, 3.8 mins elapsed, rate=41.2k reads per second ||
#> || 93% completed, 3.8 mins elapsed, rate=41.4k reads per second ||
#> || 94% completed, 3.8 mins elapsed, rate=41.6k reads per second ||
#> || 95% completed, 3.8 mins elapsed, rate=41.8k reads per second ||
#> || 96% completed, 3.9 mins elapsed, rate=41.9k reads per second ||
#> || 97% completed, 3.9 mins elapsed, rate=42.1k reads per second ||
#> || 98% completed, 3.9 mins elapsed, rate=42.3k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 10,000,000 ||
#> || Mapped : 7,118,288 (71.2%) ||
#> || Uniquely mapped : 7,118,288 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 2,881,712 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 4.2 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.5M_AC_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.5M_AC_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:46:24, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=101.1k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=109.5k reads per second ||
#> || 12% completed, 1.3 mins elapsed, rate=109.3k reads per second ||
#> || 19% completed, 1.4 mins elapsed, rate=109.4k reads per second ||
#> || 26% completed, 1.5 mins elapsed, rate=108.3k reads per second ||
#> || 32% completed, 1.7 mins elapsed, rate=107.6k reads per second ||
#> || 39% completed, 1.8 mins elapsed, rate=107.8k reads per second ||
#> || 46% completed, 1.9 mins elapsed, rate=108.0k reads per second ||
#> || 48% completed, 1.9 mins elapsed, rate=42.5k reads per second ||
#> || 51% completed, 2.0 mins elapsed, rate=43.3k reads per second ||
#> || 53% completed, 2.0 mins elapsed, rate=44.1k reads per second ||
#> || 55% completed, 2.1 mins elapsed, rate=44.8k reads per second ||
#> || 58% completed, 2.1 mins elapsed, rate=45.6k reads per second ||
#> || 60% completed, 2.2 mins elapsed, rate=46.2k reads per second ||
#> || 62% completed, 2.2 mins elapsed, rate=46.9k reads per second ||
#> || 65% completed, 2.3 mins elapsed, rate=47.5k reads per second ||
#> || 67% completed, 2.3 mins elapsed, rate=48.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 2.6 mins elapsed, rate=79.6k reads per second ||
#> || 76% completed, 2.8 mins elapsed, rate=77.6k reads per second ||
#> || 83% completed, 2.9 mins elapsed, rate=78.8k reads per second ||
#> || 90% completed, 3.0 mins elapsed, rate=49.8k reads per second ||
#> || 91% completed, 3.1 mins elapsed, rate=50.0k reads per second ||
#> || 92% completed, 3.1 mins elapsed, rate=50.2k reads per second ||
#> || 93% completed, 3.1 mins elapsed, rate=50.4k reads per second ||
#> || 94% completed, 3.1 mins elapsed, rate=50.5k reads per second ||
#> || 95% completed, 3.2 mins elapsed, rate=50.6k reads per second ||
#> || 96% completed, 3.2 mins elapsed, rate=50.8k reads per second ||
#> || 97% completed, 3.2 mins elapsed, rate=51.0k reads per second ||
#> || 98% completed, 3.2 mins elapsed, rate=51.2k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 10,000,000 ||
#> || Mapped : 7,116,281 (71.2%) ||
#> || Uniquely mapped : 7,116,281 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 2,883,719 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 3.5 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.5M_AT_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.5M_AT_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:49:55, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=99.9k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=109.7k reads per second ||
#> || 12% completed, 1.3 mins elapsed, rate=109.4k reads per second ||
#> || 19% completed, 1.4 mins elapsed, rate=109.2k reads per second ||
#> || 26% completed, 1.5 mins elapsed, rate=109.2k reads per second ||
#> || 32% completed, 1.6 mins elapsed, rate=109.2k reads per second ||
#> || 39% completed, 1.7 mins elapsed, rate=109.3k reads per second ||
#> || 46% completed, 1.8 mins elapsed, rate=109.3k reads per second ||
#> || 48% completed, 1.9 mins elapsed, rate=42.7k reads per second ||
#> || 51% completed, 2.0 mins elapsed, rate=43.5k reads per second ||
#> || 53% completed, 2.0 mins elapsed, rate=44.2k reads per second ||
#> || 55% completed, 2.1 mins elapsed, rate=45.0k reads per second ||
#> || 58% completed, 2.1 mins elapsed, rate=45.7k reads per second ||
#> || 60% completed, 2.2 mins elapsed, rate=46.4k reads per second ||
#> || 62% completed, 2.2 mins elapsed, rate=47.0k reads per second ||
#> || 65% completed, 2.3 mins elapsed, rate=47.6k reads per second ||
#> || 67% completed, 2.3 mins elapsed, rate=48.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 2.5 mins elapsed, rate=85.3k reads per second ||
#> || 76% completed, 2.6 mins elapsed, rate=87.1k reads per second ||
#> || 83% completed, 2.7 mins elapsed, rate=88.7k reads per second ||
#> || 90% completed, 2.8 mins elapsed, rate=53.5k reads per second ||
#> || 91% completed, 2.9 mins elapsed, rate=53.7k reads per second ||
#> || 92% completed, 2.9 mins elapsed, rate=53.8k reads per second ||
#> || 93% completed, 2.9 mins elapsed, rate=54.0k reads per second ||
#> || 94% completed, 2.9 mins elapsed, rate=54.1k reads per second ||
#> || 95% completed, 2.9 mins elapsed, rate=54.3k reads per second ||
#> || 96% completed, 3.0 mins elapsed, rate=54.5k reads per second ||
#> || 97% completed, 3.0 mins elapsed, rate=54.6k reads per second ||
#> || 99% completed, 3.0 mins elapsed, rate=54.7k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 10,000,000 ||
#> || Mapped : 7,116,371 (71.2%) ||
#> || Uniquely mapped : 7,116,371 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 2,883,629 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 3.3 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.5M_CA_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.5M_CA_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:53:12, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=103.8k reads per second ||
#> || 6% completed, 1.3 mins elapsed, rate=116.2k reads per second ||
#> || 13% completed, 1.4 mins elapsed, rate=116.4k reads per second ||
#> || 20% completed, 1.5 mins elapsed, rate=116.3k reads per second ||
#> || 26% completed, 1.6 mins elapsed, rate=116.3k reads per second ||
#> || 33% completed, 1.7 mins elapsed, rate=116.2k reads per second ||
#> || 40% completed, 1.8 mins elapsed, rate=116.1k reads per second ||
#> || 48% completed, 1.9 mins elapsed, rate=42.2k reads per second ||
#> || 51% completed, 2.0 mins elapsed, rate=42.8k reads per second ||
#> || 53% completed, 2.1 mins elapsed, rate=43.4k reads per second ||
#> || 55% completed, 2.1 mins elapsed, rate=44.0k reads per second ||
#> || 58% completed, 2.2 mins elapsed, rate=44.5k reads per second ||
#> || 60% completed, 2.2 mins elapsed, rate=45.0k reads per second ||
#> || 62% completed, 2.3 mins elapsed, rate=45.4k reads per second ||
#> || 65% completed, 2.4 mins elapsed, rate=45.9k reads per second ||
#> || 67% completed, 2.4 mins elapsed, rate=46.3k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 2.6 mins elapsed, rate=84.4k reads per second ||
#> || 76% completed, 2.7 mins elapsed, rate=86.4k reads per second ||
#> || 83% completed, 2.8 mins elapsed, rate=88.4k reads per second ||
#> || 90% completed, 2.9 mins elapsed, rate=52.5k reads per second ||
#> || 91% completed, 2.9 mins elapsed, rate=52.6k reads per second ||
#> || 92% completed, 2.9 mins elapsed, rate=52.7k reads per second ||
#> || 93% completed, 3.0 mins elapsed, rate=52.8k reads per second ||
#> || 94% completed, 3.0 mins elapsed, rate=52.8k reads per second ||
#> || 95% completed, 3.0 mins elapsed, rate=52.9k reads per second ||
#> || 96% completed, 3.0 mins elapsed, rate=53.0k reads per second ||
#> || 97% completed, 3.1 mins elapsed, rate=53.1k reads per second ||
#> || 98% completed, 3.1 mins elapsed, rate=53.2k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 10,000,000 ||
#> || Mapped : 1,873,121 (18.7%) ||
#> || Uniquely mapped : 1,873,121 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 8,126,879 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 3.4 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.5M_CC_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.5M_CC_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:56:33, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=103.0k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=115.5k reads per second ||
#> || 13% completed, 1.3 mins elapsed, rate=115.5k reads per second ||
#> || 19% completed, 1.4 mins elapsed, rate=114.3k reads per second ||
#> || 26% completed, 1.5 mins elapsed, rate=114.6k reads per second ||
#> || 33% completed, 1.6 mins elapsed, rate=114.8k reads per second ||
#> || 39% completed, 1.7 mins elapsed, rate=115.0k reads per second ||
#> || 46% completed, 1.8 mins elapsed, rate=114.9k reads per second ||
#> || 48% completed, 1.9 mins elapsed, rate=43.2k reads per second ||
#> || 51% completed, 2.0 mins elapsed, rate=43.8k reads per second ||
#> || 53% completed, 2.0 mins elapsed, rate=44.3k reads per second ||
#> || 55% completed, 2.1 mins elapsed, rate=44.9k reads per second ||
#> || 58% completed, 2.1 mins elapsed, rate=45.4k reads per second ||
#> || 60% completed, 2.2 mins elapsed, rate=45.9k reads per second ||
#> || 62% completed, 2.3 mins elapsed, rate=46.3k reads per second ||
#> || 65% completed, 2.3 mins elapsed, rate=46.7k reads per second ||
#> || 67% completed, 2.4 mins elapsed, rate=47.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 2.5 mins elapsed, rate=84.4k reads per second ||
#> || 76% completed, 2.6 mins elapsed, rate=86.5k reads per second ||
#> || 83% completed, 2.7 mins elapsed, rate=88.4k reads per second ||
#> || 90% completed, 2.8 mins elapsed, rate=53.4k reads per second ||
#> || 91% completed, 2.9 mins elapsed, rate=53.5k reads per second ||
#> || 92% completed, 2.9 mins elapsed, rate=53.5k reads per second ||
#> || 93% completed, 2.9 mins elapsed, rate=53.6k reads per second ||
#> || 94% completed, 2.9 mins elapsed, rate=53.7k reads per second ||
#> || 95% completed, 3.0 mins elapsed, rate=53.8k reads per second ||
#> || 96% completed, 3.0 mins elapsed, rate=53.9k reads per second ||
#> || 97% completed, 3.0 mins elapsed, rate=53.9k reads per second ||
#> || 99% completed, 3.1 mins elapsed, rate=54.0k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 10,000,000 ||
#> || Mapped : 1,876,176 (18.8%) ||
#> || Uniquely mapped : 1,876,176 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 8,123,824 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 3.3 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.5M_CT_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.5M_CT_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 17:59:52, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=104.0k reads per second ||
#> || 6% completed, 1.2 mins elapsed, rate=116.0k reads per second ||
#> || 13% completed, 1.3 mins elapsed, rate=115.9k reads per second ||
#> || 20% completed, 1.4 mins elapsed, rate=115.8k reads per second ||
#> || 26% completed, 1.5 mins elapsed, rate=115.8k reads per second ||
#> || 33% completed, 1.6 mins elapsed, rate=115.7k reads per second ||
#> || 40% completed, 1.7 mins elapsed, rate=115.8k reads per second ||
#> || 48% completed, 1.9 mins elapsed, rate=43.4k reads per second ||
#> || 51% completed, 1.9 mins elapsed, rate=43.9k reads per second ||
#> || 53% completed, 2.0 mins elapsed, rate=44.5k reads per second ||
#> || 55% completed, 2.1 mins elapsed, rate=45.1k reads per second ||
#> || 58% completed, 2.1 mins elapsed, rate=45.6k reads per second ||
#> || 60% completed, 2.2 mins elapsed, rate=46.0k reads per second ||
#> || 62% completed, 2.3 mins elapsed, rate=46.5k reads per second ||
#> || 65% completed, 2.3 mins elapsed, rate=46.9k reads per second ||
#> || 67% completed, 2.4 mins elapsed, rate=47.3k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 2.5 mins elapsed, rate=84.9k reads per second ||
#> || 76% completed, 2.6 mins elapsed, rate=87.0k reads per second ||
#> || 83% completed, 2.7 mins elapsed, rate=88.9k reads per second ||
#> || 90% completed, 2.8 mins elapsed, rate=53.6k reads per second ||
#> || 91% completed, 2.9 mins elapsed, rate=53.7k reads per second ||
#> || 92% completed, 2.9 mins elapsed, rate=53.8k reads per second ||
#> || 93% completed, 2.9 mins elapsed, rate=53.8k reads per second ||
#> || 94% completed, 2.9 mins elapsed, rate=53.9k reads per second ||
#> || 95% completed, 3.0 mins elapsed, rate=54.0k reads per second ||
#> || 96% completed, 3.0 mins elapsed, rate=54.1k reads per second ||
#> || 97% completed, 3.0 mins elapsed, rate=54.2k reads per second ||
#> || 98% completed, 3.0 mins elapsed, rate=54.2k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 10,000,000 ||
#> || Mapped : 1,876,608 (18.8%) ||
#> || Uniquely mapped : 1,876,608 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 8,123,392 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 3.3 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.10M_AA_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.10M_AA_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 18:03:25, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=96.2k reads per second ||
#> || 6% completed, 1.3 mins elapsed, rate=109.1k reads per second ||
#> || 12% completed, 1.5 mins elapsed, rate=109.2k reads per second ||
#> || 19% completed, 1.7 mins elapsed, rate=109.1k reads per second ||
#> || 24% completed, 1.9 mins elapsed, rate=42.6k reads per second ||
#> || 25% completed, 2.0 mins elapsed, rate=43.4k reads per second ||
#> || 26% completed, 2.0 mins elapsed, rate=44.2k reads per second ||
#> || 27% completed, 2.1 mins elapsed, rate=44.9k reads per second ||
#> || 29% completed, 2.1 mins elapsed, rate=45.6k reads per second ||
#> || 30% completed, 2.2 mins elapsed, rate=46.3k reads per second ||
#> || 31% completed, 2.2 mins elapsed, rate=47.0k reads per second ||
#> || 32% completed, 2.3 mins elapsed, rate=47.6k reads per second ||
#> || 33% completed, 2.3 mins elapsed, rate=48.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 2.5 mins elapsed, rate=84.4k reads per second ||
#> || 41% completed, 2.7 mins elapsed, rate=87.8k reads per second ||
#> || 48% completed, 2.9 mins elapsed, rate=90.6k reads per second ||
#> || 54% completed, 3.1 mins elapsed, rate=92.8k reads per second ||
#> || 59% completed, 3.3 mins elapsed, rate=60.6k reads per second ||
#> || 60% completed, 3.3 mins elapsed, rate=60.8k reads per second ||
#> || 61% completed, 3.4 mins elapsed, rate=61.0k reads per second ||
#> || 62% completed, 3.4 mins elapsed, rate=61.2k reads per second ||
#> || 64% completed, 3.5 mins elapsed, rate=61.4k reads per second ||
#> || 65% completed, 3.5 mins elapsed, rate=61.6k reads per second ||
#> || 66% completed, 3.6 mins elapsed, rate=61.8k reads per second ||
#> || 67% completed, 3.6 mins elapsed, rate=62.0k reads per second ||
#> || 68% completed, 3.7 mins elapsed, rate=62.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 3.9 mins elapsed, rate=85.9k reads per second ||
#> || 76% completed, 4.1 mins elapsed, rate=87.7k reads per second ||
#> || 83% completed, 4.3 mins elapsed, rate=89.3k reads per second ||
#> || 89% completed, 4.5 mins elapsed, rate=90.7k reads per second ||
#> || 90% completed, 4.5 mins elapsed, rate=67.4k reads per second ||
#> || 91% completed, 4.5 mins elapsed, rate=67.4k reads per second ||
#> || 92% completed, 4.6 mins elapsed, rate=67.5k reads per second ||
#> || 93% completed, 4.6 mins elapsed, rate=67.6k reads per second ||
#> || 94% completed, 4.7 mins elapsed, rate=67.7k reads per second ||
#> || 95% completed, 4.7 mins elapsed, rate=67.7k reads per second ||
#> || 96% completed, 4.8 mins elapsed, rate=67.8k reads per second ||
#> || 97% completed, 4.8 mins elapsed, rate=67.9k reads per second ||
#> || 98% completed, 4.9 mins elapsed, rate=67.9k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 20,000,000 ||
#> || Mapped : 14,236,795 (71.2%) ||
#> || Uniquely mapped : 14,236,795 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 5,763,205 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 5.4 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.10M_AC_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.10M_AC_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 18:08:51, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=100.2k reads per second ||
#> || 6% completed, 1.3 mins elapsed, rate=109.1k reads per second ||
#> || 12% completed, 1.5 mins elapsed, rate=109.2k reads per second ||
#> || 19% completed, 1.7 mins elapsed, rate=109.2k reads per second ||
#> || 24% completed, 1.9 mins elapsed, rate=42.6k reads per second ||
#> || 25% completed, 2.0 mins elapsed, rate=43.4k reads per second ||
#> || 26% completed, 2.0 mins elapsed, rate=44.2k reads per second ||
#> || 27% completed, 2.1 mins elapsed, rate=44.9k reads per second ||
#> || 29% completed, 2.1 mins elapsed, rate=45.6k reads per second ||
#> || 30% completed, 2.2 mins elapsed, rate=46.3k reads per second ||
#> || 31% completed, 2.2 mins elapsed, rate=46.9k reads per second ||
#> || 32% completed, 2.3 mins elapsed, rate=47.5k reads per second ||
#> || 33% completed, 2.3 mins elapsed, rate=48.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 2.5 mins elapsed, rate=85.5k reads per second ||
#> || 41% completed, 2.7 mins elapsed, rate=88.9k reads per second ||
#> || 48% completed, 2.9 mins elapsed, rate=91.5k reads per second ||
#> || 54% completed, 3.1 mins elapsed, rate=93.7k reads per second ||
#> || 59% completed, 3.3 mins elapsed, rate=60.8k reads per second ||
#> || 60% completed, 3.3 mins elapsed, rate=61.0k reads per second ||
#> || 61% completed, 3.4 mins elapsed, rate=61.2k reads per second ||
#> || 62% completed, 3.4 mins elapsed, rate=61.4k reads per second ||
#> || 63% completed, 3.5 mins elapsed, rate=61.7k reads per second ||
#> || 65% completed, 3.5 mins elapsed, rate=61.9k reads per second ||
#> || 66% completed, 3.6 mins elapsed, rate=62.1k reads per second ||
#> || 67% completed, 3.6 mins elapsed, rate=62.2k reads per second ||
#> || 68% completed, 3.7 mins elapsed, rate=62.4k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 3.8 mins elapsed, rate=86.4k reads per second ||
#> || 76% completed, 4.0 mins elapsed, rate=88.2k reads per second ||
#> || 83% completed, 4.2 mins elapsed, rate=89.7k reads per second ||
#> || 89% completed, 4.4 mins elapsed, rate=90.9k reads per second ||
#> || 90% completed, 4.5 mins elapsed, rate=67.4k reads per second ||
#> || 91% completed, 4.5 mins elapsed, rate=67.5k reads per second ||
#> || 92% completed, 4.6 mins elapsed, rate=67.6k reads per second ||
#> || 93% completed, 4.6 mins elapsed, rate=67.6k reads per second ||
#> || 94% completed, 4.7 mins elapsed, rate=67.7k reads per second ||
#> || 95% completed, 4.7 mins elapsed, rate=67.8k reads per second ||
#> || 96% completed, 4.8 mins elapsed, rate=67.9k reads per second ||
#> || 97% completed, 4.8 mins elapsed, rate=67.9k reads per second ||
#> || 98% completed, 4.9 mins elapsed, rate=68.0k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 20,000,000 ||
#> || Mapped : 14,233,458 (71.2%) ||
#> || Uniquely mapped : 14,233,458 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 5,766,542 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 5.4 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.10M_AT_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.10M_AT_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 18:14:16, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=100.2k reads per second ||
#> || 6% completed, 1.3 mins elapsed, rate=109.2k reads per second ||
#> || 12% completed, 1.5 mins elapsed, rate=109.2k reads per second ||
#> || 19% completed, 1.7 mins elapsed, rate=109.1k reads per second ||
#> || 24% completed, 1.9 mins elapsed, rate=42.6k reads per second ||
#> || 25% completed, 2.0 mins elapsed, rate=43.4k reads per second ||
#> || 26% completed, 2.0 mins elapsed, rate=44.2k reads per second ||
#> || 27% completed, 2.1 mins elapsed, rate=44.9k reads per second ||
#> || 29% completed, 2.1 mins elapsed, rate=45.7k reads per second ||
#> || 30% completed, 2.2 mins elapsed, rate=46.3k reads per second ||
#> || 31% completed, 2.2 mins elapsed, rate=47.0k reads per second ||
#> || 32% completed, 2.3 mins elapsed, rate=47.6k reads per second ||
#> || 33% completed, 2.3 mins elapsed, rate=48.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 2.5 mins elapsed, rate=84.4k reads per second ||
#> || 41% completed, 2.7 mins elapsed, rate=87.9k reads per second ||
#> || 48% completed, 2.9 mins elapsed, rate=90.7k reads per second ||
#> || 54% completed, 3.1 mins elapsed, rate=92.9k reads per second ||
#> || 59% completed, 3.3 mins elapsed, rate=60.5k reads per second ||
#> || 60% completed, 3.3 mins elapsed, rate=60.8k reads per second ||
#> || 61% completed, 3.4 mins elapsed, rate=61.0k reads per second ||
#> || 62% completed, 3.4 mins elapsed, rate=61.2k reads per second ||
#> || 63% completed, 3.5 mins elapsed, rate=61.4k reads per second ||
#> || 65% completed, 3.5 mins elapsed, rate=61.6k reads per second ||
#> || 66% completed, 3.6 mins elapsed, rate=61.8k reads per second ||
#> || 67% completed, 3.6 mins elapsed, rate=62.0k reads per second ||
#> || 68% completed, 3.7 mins elapsed, rate=62.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 3.9 mins elapsed, rate=86.0k reads per second ||
#> || 76% completed, 4.1 mins elapsed, rate=87.8k reads per second ||
#> || 83% completed, 4.3 mins elapsed, rate=89.4k reads per second ||
#> || 89% completed, 4.4 mins elapsed, rate=90.8k reads per second ||
#> || 90% completed, 4.5 mins elapsed, rate=67.4k reads per second ||
#> || 91% completed, 4.5 mins elapsed, rate=67.5k reads per second ||
#> || 92% completed, 4.6 mins elapsed, rate=67.6k reads per second ||
#> || 93% completed, 4.6 mins elapsed, rate=67.6k reads per second ||
#> || 94% completed, 4.7 mins elapsed, rate=67.7k reads per second ||
#> || 95% completed, 4.7 mins elapsed, rate=67.8k reads per second ||
#> || 96% completed, 4.8 mins elapsed, rate=67.8k reads per second ||
#> || 97% completed, 4.8 mins elapsed, rate=67.9k reads per second ||
#> || 98% completed, 4.9 mins elapsed, rate=68.0k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 20,000,000 ||
#> || Mapped : 14,234,627 (71.2%) ||
#> || Uniquely mapped : 14,234,627 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 5,765,373 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 5.4 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.10M_CA_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.10M_CA_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 18:19:40, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=102.8k reads per second ||
#> || 6% completed, 1.4 mins elapsed, rate=111.8k reads per second ||
#> || 13% completed, 1.6 mins elapsed, rate=111.9k reads per second ||
#> || 20% completed, 1.8 mins elapsed, rate=112.2k reads per second ||
#> || 24% completed, 1.9 mins elapsed, rate=42.4k reads per second ||
#> || 25% completed, 2.0 mins elapsed, rate=43.0k reads per second ||
#> || 26% completed, 2.1 mins elapsed, rate=43.5k reads per second ||
#> || 27% completed, 2.1 mins elapsed, rate=44.0k reads per second ||
#> || 29% completed, 2.2 mins elapsed, rate=44.5k reads per second ||
#> || 30% completed, 2.2 mins elapsed, rate=45.0k reads per second ||
#> || 31% completed, 2.3 mins elapsed, rate=45.4k reads per second ||
#> || 32% completed, 2.4 mins elapsed, rate=45.9k reads per second ||
#> || 33% completed, 2.4 mins elapsed, rate=46.3k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 2.6 mins elapsed, rate=81.5k reads per second ||
#> || 41% completed, 2.8 mins elapsed, rate=85.6k reads per second ||
#> || 48% completed, 3.0 mins elapsed, rate=88.9k reads per second ||
#> || 54% completed, 3.2 mins elapsed, rate=91.5k reads per second ||
#> || 59% completed, 3.3 mins elapsed, rate=59.4k reads per second ||
#> || 60% completed, 3.4 mins elapsed, rate=59.5k reads per second ||
#> || 61% completed, 3.5 mins elapsed, rate=59.5k reads per second ||
#> || 62% completed, 3.5 mins elapsed, rate=59.6k reads per second ||
#> || 64% completed, 3.6 mins elapsed, rate=59.6k reads per second ||
#> || 65% completed, 3.6 mins elapsed, rate=59.6k reads per second ||
#> || 66% completed, 3.7 mins elapsed, rate=59.7k reads per second ||
#> || 67% completed, 3.8 mins elapsed, rate=59.7k reads per second ||
#> || 68% completed, 3.8 mins elapsed, rate=59.8k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 4.0 mins elapsed, rate=83.2k reads per second ||
#> || 76% completed, 4.2 mins elapsed, rate=85.3k reads per second ||
#> || 83% completed, 4.4 mins elapsed, rate=87.1k reads per second ||
#> || 89% completed, 4.5 mins elapsed, rate=88.7k reads per second ||
#> || 90% completed, 4.6 mins elapsed, rate=65.9k reads per second ||
#> || 91% completed, 4.7 mins elapsed, rate=65.8k reads per second ||
#> || 92% completed, 4.7 mins elapsed, rate=65.8k reads per second ||
#> || 93% completed, 4.8 mins elapsed, rate=65.8k reads per second ||
#> || 94% completed, 4.8 mins elapsed, rate=65.7k reads per second ||
#> || 96% completed, 4.9 mins elapsed, rate=65.7k reads per second ||
#> || 97% completed, 4.9 mins elapsed, rate=65.6k reads per second ||
#> || 98% completed, 5.0 mins elapsed, rate=65.6k reads per second ||
#> || 99% completed, 5.0 mins elapsed, rate=65.6k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 20,000,000 ||
#> || Mapped : 3,747,824 (18.7%) ||
#> || Uniquely mapped : 3,747,824 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 16,252,176 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 5.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.10M_CC_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.10M_CC_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 18:25:17, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=103.1k reads per second ||
#> || 6% completed, 1.4 mins elapsed, rate=113.0k reads per second ||
#> || 13% completed, 1.6 mins elapsed, rate=112.8k reads per second ||
#> || 19% completed, 1.7 mins elapsed, rate=112.5k reads per second ||
#> || 24% completed, 1.9 mins elapsed, rate=42.5k reads per second ||
#> || 25% completed, 2.0 mins elapsed, rate=43.1k reads per second ||
#> || 26% completed, 2.0 mins elapsed, rate=43.6k reads per second ||
#> || 27% completed, 2.1 mins elapsed, rate=44.1k reads per second ||
#> || 29% completed, 2.2 mins elapsed, rate=44.6k reads per second ||
#> || 30% completed, 2.2 mins elapsed, rate=45.1k reads per second ||
#> || 31% completed, 2.3 mins elapsed, rate=45.5k reads per second ||
#> || 32% completed, 2.4 mins elapsed, rate=45.9k reads per second ||
#> || 33% completed, 2.4 mins elapsed, rate=46.3k reads per second ||
#> || Start read mapping in chunk. ||
#> || 35% completed, 2.6 mins elapsed, rate=82.2k reads per second ||
#> || 41% completed, 2.8 mins elapsed, rate=86.0k reads per second ||
#> || 48% completed, 3.0 mins elapsed, rate=89.2k reads per second ||
#> || 55% completed, 3.2 mins elapsed, rate=91.8k reads per second ||
#> || 59% completed, 3.3 mins elapsed, rate=59.7k reads per second ||
#> || 60% completed, 3.4 mins elapsed, rate=59.8k reads per second ||
#> || 61% completed, 3.4 mins elapsed, rate=59.8k reads per second ||
#> || 62% completed, 3.5 mins elapsed, rate=59.9k reads per second ||
#> || 64% completed, 3.6 mins elapsed, rate=59.9k reads per second ||
#> || 65% completed, 3.6 mins elapsed, rate=60.0k reads per second ||
#> || 66% completed, 3.7 mins elapsed, rate=60.0k reads per second ||
#> || 67% completed, 3.8 mins elapsed, rate=60.0k reads per second ||
#> || 68% completed, 3.8 mins elapsed, rate=60.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 4.0 mins elapsed, rate=83.4k reads per second ||
#> || 76% completed, 4.1 mins elapsed, rate=85.5k reads per second ||
#> || 83% completed, 4.3 mins elapsed, rate=87.4k reads per second ||
#> || 89% completed, 4.5 mins elapsed, rate=89.1k reads per second ||
#> || 90% completed, 4.6 mins elapsed, rate=66.1k reads per second ||
#> || 91% completed, 4.6 mins elapsed, rate=66.1k reads per second ||
#> || 92% completed, 4.7 mins elapsed, rate=66.0k reads per second ||
#> || 93% completed, 4.7 mins elapsed, rate=66.0k reads per second ||
#> || 94% completed, 4.8 mins elapsed, rate=65.9k reads per second ||
#> || 95% completed, 4.9 mins elapsed, rate=65.9k reads per second ||
#> || 96% completed, 4.9 mins elapsed, rate=65.8k reads per second ||
#> || 97% completed, 5.0 mins elapsed, rate=65.8k reads per second ||
#> || 98% completed, 5.0 mins elapsed, rate=65.7k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 20,000,000 ||
#> || Mapped : 3,751,602 (18.8%) ||
#> || Uniquely mapped : 3,751,602 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 16,248,398 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 5.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.10M_CT_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.10M_CT_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 18:30:52, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=101.1k reads per second ||
#> || 6% completed, 1.4 mins elapsed, rate=112.6k reads per second ||
#> || 13% completed, 1.6 mins elapsed, rate=112.5k reads per second ||
#> || 20% completed, 1.8 mins elapsed, rate=112.5k reads per second ||
#> || 24% completed, 1.9 mins elapsed, rate=42.5k reads per second ||
#> || 25% completed, 2.0 mins elapsed, rate=43.1k reads per second ||
#> || 26% completed, 2.0 mins elapsed, rate=43.6k reads per second ||
#> || 27% completed, 2.1 mins elapsed, rate=44.1k reads per second ||
#> || 28% completed, 2.2 mins elapsed, rate=44.6k reads per second ||
#> || 30% completed, 2.2 mins elapsed, rate=45.1k reads per second ||
#> || 31% completed, 2.3 mins elapsed, rate=45.5k reads per second ||
#> || 32% completed, 2.4 mins elapsed, rate=45.8k reads per second ||
#> || 33% completed, 2.4 mins elapsed, rate=46.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 2.6 mins elapsed, rate=82.0k reads per second ||
#> || 41% completed, 2.8 mins elapsed, rate=86.0k reads per second ||
#> || 48% completed, 3.0 mins elapsed, rate=89.3k reads per second ||
#> || 54% completed, 3.2 mins elapsed, rate=91.9k reads per second ||
#> || 59% completed, 3.3 mins elapsed, rate=59.6k reads per second ||
#> || 60% completed, 3.4 mins elapsed, rate=59.7k reads per second ||
#> || 61% completed, 3.4 mins elapsed, rate=59.7k reads per second ||
#> || 62% completed, 3.5 mins elapsed, rate=59.8k reads per second ||
#> || 63% completed, 3.6 mins elapsed, rate=59.8k reads per second ||
#> || 65% completed, 3.6 mins elapsed, rate=59.9k reads per second ||
#> || 66% completed, 3.7 mins elapsed, rate=59.9k reads per second ||
#> || 67% completed, 3.8 mins elapsed, rate=59.9k reads per second ||
#> || 68% completed, 3.8 mins elapsed, rate=60.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 4.0 mins elapsed, rate=83.6k reads per second ||
#> || 76% completed, 4.1 mins elapsed, rate=85.7k reads per second ||
#> || 83% completed, 4.3 mins elapsed, rate=87.5k reads per second ||
#> || 89% completed, 4.5 mins elapsed, rate=89.2k reads per second ||
#> || 90% completed, 4.6 mins elapsed, rate=66.1k reads per second ||
#> || 91% completed, 4.6 mins elapsed, rate=66.1k reads per second ||
#> || 92% completed, 4.7 mins elapsed, rate=66.0k reads per second ||
#> || 93% completed, 4.7 mins elapsed, rate=66.0k reads per second ||
#> || 94% completed, 4.8 mins elapsed, rate=65.9k reads per second ||
#> || 95% completed, 4.9 mins elapsed, rate=65.9k reads per second ||
#> || 96% completed, 4.9 mins elapsed, rate=65.9k reads per second ||
#> || 97% completed, 5.0 mins elapsed, rate=65.8k reads per second ||
#> || 98% completed, 5.0 mins elapsed, rate=65.8k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 20,000,000 ||
#> || Mapped : 3,750,703 (18.8%) ||
#> || Uniquely mapped : 3,750,703 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 16,249,297 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 5.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.15M_AA_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.15M_AA_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 18:36:31, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=96.8k reads per second ||
#> || 6% completed, 1.4 mins elapsed, rate=106.3k reads per second ||
#> || 12% completed, 1.8 mins elapsed, rate=106.7k reads per second ||
#> || 16% completed, 1.9 mins elapsed, rate=41.9k reads per second ||
#> || 17% completed, 2.0 mins elapsed, rate=42.7k reads per second ||
#> || 17% completed, 2.1 mins elapsed, rate=43.5k reads per second ||
#> || 18% completed, 2.1 mins elapsed, rate=44.2k reads per second ||
#> || 19% completed, 2.2 mins elapsed, rate=44.9k reads per second ||
#> || 20% completed, 2.2 mins elapsed, rate=45.6k reads per second ||
#> || 20% completed, 2.3 mins elapsed, rate=46.2k reads per second ||
#> || 21% completed, 2.3 mins elapsed, rate=46.8k reads per second ||
#> || 22% completed, 2.4 mins elapsed, rate=47.4k reads per second ||
#> || Start read mapping in chunk. ||
#> || 23% completed, 2.6 mins elapsed, rate=83.4k reads per second ||
#> || 29% completed, 2.9 mins elapsed, rate=88.3k reads per second ||
#> || 36% completed, 3.2 mins elapsed, rate=91.6k reads per second ||
#> || 39% completed, 3.3 mins elapsed, rate=59.7k reads per second ||
#> || 40% completed, 3.4 mins elapsed, rate=59.9k reads per second ||
#> || 41% completed, 3.4 mins elapsed, rate=60.1k reads per second ||
#> || 41% completed, 3.5 mins elapsed, rate=60.4k reads per second ||
#> || 42% completed, 3.5 mins elapsed, rate=60.5k reads per second ||
#> || 43% completed, 3.6 mins elapsed, rate=60.8k reads per second ||
#> || 44% completed, 3.6 mins elapsed, rate=60.9k reads per second ||
#> || 44% completed, 3.7 mins elapsed, rate=61.1k reads per second ||
#> || 45% completed, 3.7 mins elapsed, rate=61.3k reads per second ||
#> || Start read mapping in chunk. ||
#> || 46% completed, 3.9 mins elapsed, rate=84.9k reads per second ||
#> || 53% completed, 4.2 mins elapsed, rate=87.4k reads per second ||
#> || 59% completed, 4.5 mins elapsed, rate=89.4k reads per second ||
#> || 62% completed, 4.7 mins elapsed, rate=67.4k reads per second ||
#> || 63% completed, 4.7 mins elapsed, rate=67.5k reads per second ||
#> || 64% completed, 4.8 mins elapsed, rate=67.5k reads per second ||
#> || 65% completed, 4.8 mins elapsed, rate=67.6k reads per second ||
#> || 65% completed, 4.9 mins elapsed, rate=67.7k reads per second ||
#> || 66% completed, 4.9 mins elapsed, rate=67.8k reads per second ||
#> || 67% completed, 5.0 mins elapsed, rate=67.8k reads per second ||
#> || 68% completed, 5.0 mins elapsed, rate=67.9k reads per second ||
#> || 69% completed, 5.1 mins elapsed, rate=68.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 5.3 mins elapsed, rate=85.1k reads per second ||
#> || 76% completed, 5.6 mins elapsed, rate=86.9k reads per second ||
#> || 83% completed, 5.9 mins elapsed, rate=88.4k reads per second ||
#> || 86% completed, 6.0 mins elapsed, rate=71.5k reads per second ||
#> || 86% completed, 6.1 mins elapsed, rate=71.6k reads per second ||
#> || 87% completed, 6.1 mins elapsed, rate=71.6k reads per second ||
#> || 88% completed, 6.2 mins elapsed, rate=71.6k reads per second ||
#> || 89% completed, 6.2 mins elapsed, rate=71.6k reads per second ||
#> || 89% completed, 6.3 mins elapsed, rate=71.6k reads per second ||
#> || 90% completed, 6.3 mins elapsed, rate=71.6k reads per second ||
#> || 91% completed, 6.4 mins elapsed, rate=71.7k reads per second ||
#> || 92% completed, 6.4 mins elapsed, rate=71.7k reads per second ||
#> || Start read mapping in chunk. ||
#> || 93% completed, 6.6 mins elapsed, rate=85.4k reads per second ||
#> || 97% completed, 6.8 mins elapsed, rate=71.6k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=71.6k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=71.6k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=71.6k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=71.6k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=71.6k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=71.6k reads per second ||
#> || 99% completed, 7.0 mins elapsed, rate=71.6k reads per second ||
#> || 99% completed, 7.0 mins elapsed, rate=71.6k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 30,000,000 ||
#> || Mapped : 21,355,573 (71.2%) ||
#> || Uniquely mapped : 21,355,573 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 8,644,427 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 7.7 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.15M_AC_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.15M_AC_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 18:44:13, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=101.8k reads per second ||
#> || 6% completed, 1.5 mins elapsed, rate=106.9k reads per second ||
#> || 12% completed, 1.8 mins elapsed, rate=107.0k reads per second ||
#> || 16% completed, 1.9 mins elapsed, rate=42.0k reads per second ||
#> || 17% completed, 2.0 mins elapsed, rate=42.7k reads per second ||
#> || 17% completed, 2.1 mins elapsed, rate=43.5k reads per second ||
#> || 18% completed, 2.1 mins elapsed, rate=44.2k reads per second ||
#> || 19% completed, 2.2 mins elapsed, rate=44.9k reads per second ||
#> || 20% completed, 2.2 mins elapsed, rate=45.6k reads per second ||
#> || 20% completed, 2.3 mins elapsed, rate=46.2k reads per second ||
#> || 21% completed, 2.3 mins elapsed, rate=46.8k reads per second ||
#> || 22% completed, 2.4 mins elapsed, rate=47.4k reads per second ||
#> || Start read mapping in chunk. ||
#> || 23% completed, 2.6 mins elapsed, rate=83.6k reads per second ||
#> || 29% completed, 2.9 mins elapsed, rate=88.4k reads per second ||
#> || 36% completed, 3.2 mins elapsed, rate=91.9k reads per second ||
#> || 39% completed, 3.3 mins elapsed, rate=59.8k reads per second ||
#> || 40% completed, 3.4 mins elapsed, rate=60.0k reads per second ||
#> || 41% completed, 3.4 mins elapsed, rate=60.2k reads per second ||
#> || 41% completed, 3.5 mins elapsed, rate=60.4k reads per second ||
#> || 42% completed, 3.5 mins elapsed, rate=60.6k reads per second ||
#> || 43% completed, 3.6 mins elapsed, rate=60.8k reads per second ||
#> || 44% completed, 3.6 mins elapsed, rate=61.0k reads per second ||
#> || 44% completed, 3.7 mins elapsed, rate=61.2k reads per second ||
#> || 45% completed, 3.7 mins elapsed, rate=61.4k reads per second ||
#> || Start read mapping in chunk. ||
#> || 46% completed, 3.9 mins elapsed, rate=85.0k reads per second ||
#> || 53% completed, 4.2 mins elapsed, rate=87.5k reads per second ||
#> || 59% completed, 4.5 mins elapsed, rate=89.6k reads per second ||
#> || 62% completed, 4.7 mins elapsed, rate=67.4k reads per second ||
#> || 63% completed, 4.7 mins elapsed, rate=67.5k reads per second ||
#> || 64% completed, 4.8 mins elapsed, rate=67.6k reads per second ||
#> || 65% completed, 4.8 mins elapsed, rate=67.6k reads per second ||
#> || 65% completed, 4.9 mins elapsed, rate=67.7k reads per second ||
#> || 66% completed, 4.9 mins elapsed, rate=67.8k reads per second ||
#> || 67% completed, 5.0 mins elapsed, rate=67.8k reads per second ||
#> || 68% completed, 5.0 mins elapsed, rate=67.9k reads per second ||
#> || 69% completed, 5.1 mins elapsed, rate=68.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 5.3 mins elapsed, rate=85.4k reads per second ||
#> || 76% completed, 5.6 mins elapsed, rate=87.1k reads per second ||
#> || 83% completed, 5.9 mins elapsed, rate=88.7k reads per second ||
#> || 86% completed, 6.0 mins elapsed, rate=71.8k reads per second ||
#> || 86% completed, 6.1 mins elapsed, rate=71.8k reads per second ||
#> || 87% completed, 6.1 mins elapsed, rate=71.8k reads per second ||
#> || 88% completed, 6.2 mins elapsed, rate=71.9k reads per second ||
#> || 89% completed, 6.2 mins elapsed, rate=71.9k reads per second ||
#> || 90% completed, 6.3 mins elapsed, rate=71.9k reads per second ||
#> || 90% completed, 6.3 mins elapsed, rate=72.0k reads per second ||
#> || 91% completed, 6.4 mins elapsed, rate=72.0k reads per second ||
#> || 92% completed, 6.4 mins elapsed, rate=72.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 93% completed, 6.6 mins elapsed, rate=85.9k reads per second ||
#> || 97% completed, 6.8 mins elapsed, rate=72.0k reads per second ||
#> || 98% completed, 6.8 mins elapsed, rate=72.0k reads per second ||
#> || 98% completed, 6.8 mins elapsed, rate=72.0k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=72.0k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=72.0k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=72.0k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=72.0k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=72.0k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=72.0k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 30,000,000 ||
#> || Mapped : 21,351,406 (71.2%) ||
#> || Uniquely mapped : 21,351,406 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 8,648,594 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 7.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.15M_AT_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.15M_AT_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 18:51:50, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=75.0k reads per second ||
#> || 6% completed, 1.4 mins elapsed, rate=108.6k reads per second ||
#> || 12% completed, 1.7 mins elapsed, rate=108.9k reads per second ||
#> || 16% completed, 1.9 mins elapsed, rate=42.5k reads per second ||
#> || 17% completed, 2.0 mins elapsed, rate=43.3k reads per second ||
#> || 17% completed, 2.0 mins elapsed, rate=44.1k reads per second ||
#> || 18% completed, 2.1 mins elapsed, rate=44.8k reads per second ||
#> || 19% completed, 2.1 mins elapsed, rate=45.5k reads per second ||
#> || 20% completed, 2.2 mins elapsed, rate=46.2k reads per second ||
#> || 20% completed, 2.2 mins elapsed, rate=46.8k reads per second ||
#> || 21% completed, 2.3 mins elapsed, rate=47.4k reads per second ||
#> || 22% completed, 2.3 mins elapsed, rate=48.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 23% completed, 2.5 mins elapsed, rate=85.3k reads per second ||
#> || 29% completed, 2.8 mins elapsed, rate=90.2k reads per second ||
#> || 36% completed, 3.1 mins elapsed, rate=93.7k reads per second ||
#> || 39% completed, 3.3 mins elapsed, rate=60.7k reads per second ||
#> || 40% completed, 3.3 mins elapsed, rate=60.9k reads per second ||
#> || 41% completed, 3.4 mins elapsed, rate=61.2k reads per second ||
#> || 41% completed, 3.4 mins elapsed, rate=61.4k reads per second ||
#> || 42% completed, 3.5 mins elapsed, rate=61.6k reads per second ||
#> || 43% completed, 3.5 mins elapsed, rate=61.8k reads per second ||
#> || 44% completed, 3.6 mins elapsed, rate=62.0k reads per second ||
#> || 44% completed, 3.6 mins elapsed, rate=62.2k reads per second ||
#> || 45% completed, 3.7 mins elapsed, rate=62.4k reads per second ||
#> || Start read mapping in chunk. ||
#> || 46% completed, 3.8 mins elapsed, rate=87.0k reads per second ||
#> || 53% completed, 4.1 mins elapsed, rate=89.6k reads per second ||
#> || 59% completed, 4.4 mins elapsed, rate=91.7k reads per second ||
#> || 62% completed, 4.6 mins elapsed, rate=68.8k reads per second ||
#> || 63% completed, 4.6 mins elapsed, rate=68.9k reads per second ||
#> || 64% completed, 4.7 mins elapsed, rate=69.0k reads per second ||
#> || 65% completed, 4.7 mins elapsed, rate=69.0k reads per second ||
#> || 65% completed, 4.8 mins elapsed, rate=69.1k reads per second ||
#> || 66% completed, 4.8 mins elapsed, rate=69.2k reads per second ||
#> || 67% completed, 4.9 mins elapsed, rate=69.2k reads per second ||
#> || 68% completed, 4.9 mins elapsed, rate=69.3k reads per second ||
#> || 69% completed, 5.0 mins elapsed, rate=69.3k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 5.1 mins elapsed, rate=87.5k reads per second ||
#> || 76% completed, 5.4 mins elapsed, rate=89.2k reads per second ||
#> || 83% completed, 5.7 mins elapsed, rate=90.8k reads per second ||
#> || 86% completed, 5.9 mins elapsed, rate=73.2k reads per second ||
#> || 86% completed, 5.9 mins elapsed, rate=73.2k reads per second ||
#> || 87% completed, 6.0 mins elapsed, rate=73.2k reads per second ||
#> || 88% completed, 6.0 mins elapsed, rate=73.2k reads per second ||
#> || 89% completed, 6.1 mins elapsed, rate=73.2k reads per second ||
#> || 90% completed, 6.1 mins elapsed, rate=73.2k reads per second ||
#> || 90% completed, 6.2 mins elapsed, rate=73.2k reads per second ||
#> || 91% completed, 6.3 mins elapsed, rate=73.3k reads per second ||
#> || 92% completed, 6.3 mins elapsed, rate=73.3k reads per second ||
#> || Start read mapping in chunk. ||
#> || 93% completed, 6.5 mins elapsed, rate=87.5k reads per second ||
#> || 97% completed, 6.7 mins elapsed, rate=73.1k reads per second ||
#> || 98% completed, 6.7 mins elapsed, rate=73.1k reads per second ||
#> || 98% completed, 6.7 mins elapsed, rate=73.1k reads per second ||
#> || 98% completed, 6.7 mins elapsed, rate=73.1k reads per second ||
#> || 98% completed, 6.8 mins elapsed, rate=73.1k reads per second ||
#> || 99% completed, 6.8 mins elapsed, rate=73.1k reads per second ||
#> || 99% completed, 6.8 mins elapsed, rate=73.1k reads per second ||
#> || 99% completed, 6.8 mins elapsed, rate=73.1k reads per second ||
#> || 99% completed, 6.8 mins elapsed, rate=73.1k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 30,000,000 ||
#> || Mapped : 21,351,123 (71.2%) ||
#> || Uniquely mapped : 21,351,123 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 8,648,877 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 7.5 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.15M_CA_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.15M_CA_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 18:59:22, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=77.6k reads per second ||
#> || 6% completed, 1.4 mins elapsed, rate=115.3k reads per second ||
#> || 13% completed, 1.7 mins elapsed, rate=115.2k reads per second ||
#> || 16% completed, 1.9 mins elapsed, rate=43.2k reads per second ||
#> || 17% completed, 2.0 mins elapsed, rate=43.7k reads per second ||
#> || 17% completed, 2.0 mins elapsed, rate=44.3k reads per second ||
#> || 18% completed, 2.1 mins elapsed, rate=44.8k reads per second ||
#> || 19% completed, 2.1 mins elapsed, rate=45.3k reads per second ||
#> || 20% completed, 2.2 mins elapsed, rate=45.8k reads per second ||
#> || 20% completed, 2.3 mins elapsed, rate=46.2k reads per second ||
#> || 21% completed, 2.3 mins elapsed, rate=46.7k reads per second ||
#> || 22% completed, 2.4 mins elapsed, rate=47.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 23% completed, 2.5 mins elapsed, rate=83.6k reads per second ||
#> || 29% completed, 2.8 mins elapsed, rate=89.5k reads per second ||
#> || 36% completed, 3.1 mins elapsed, rate=93.8k reads per second ||
#> || 39% completed, 3.3 mins elapsed, rate=60.8k reads per second ||
#> || 40% completed, 3.3 mins elapsed, rate=60.8k reads per second ||
#> || 41% completed, 3.4 mins elapsed, rate=60.9k reads per second ||
#> || 41% completed, 3.4 mins elapsed, rate=60.9k reads per second ||
#> || 42% completed, 3.5 mins elapsed, rate=61.0k reads per second ||
#> || 43% completed, 3.6 mins elapsed, rate=61.0k reads per second ||
#> || 44% completed, 3.6 mins elapsed, rate=61.0k reads per second ||
#> || 45% completed, 3.7 mins elapsed, rate=61.0k reads per second ||
#> || 45% completed, 3.8 mins elapsed, rate=61.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 46% completed, 3.9 mins elapsed, rate=84.5k reads per second ||
#> || 53% completed, 4.2 mins elapsed, rate=87.7k reads per second ||
#> || 59% completed, 4.5 mins elapsed, rate=90.3k reads per second ||
#> || 62% completed, 4.6 mins elapsed, rate=68.1k reads per second ||
#> || 63% completed, 4.7 mins elapsed, rate=68.0k reads per second ||
#> || 64% completed, 4.7 mins elapsed, rate=68.0k reads per second ||
#> || 65% completed, 4.8 mins elapsed, rate=67.9k reads per second ||
#> || 66% completed, 4.9 mins elapsed, rate=67.8k reads per second ||
#> || 66% completed, 4.9 mins elapsed, rate=67.8k reads per second ||
#> || 67% completed, 5.0 mins elapsed, rate=67.7k reads per second ||
#> || 68% completed, 5.1 mins elapsed, rate=67.7k reads per second ||
#> || 69% completed, 5.1 mins elapsed, rate=67.6k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 5.3 mins elapsed, rate=85.1k reads per second ||
#> || 76% completed, 5.5 mins elapsed, rate=87.2k reads per second ||
#> || 83% completed, 5.8 mins elapsed, rate=89.1k reads per second ||
#> || 86% completed, 6.0 mins elapsed, rate=72.2k reads per second ||
#> || 86% completed, 6.0 mins elapsed, rate=72.1k reads per second ||
#> || 87% completed, 6.1 mins elapsed, rate=72.0k reads per second ||
#> || 88% completed, 6.2 mins elapsed, rate=71.9k reads per second ||
#> || 89% completed, 6.2 mins elapsed, rate=71.8k reads per second ||
#> || 90% completed, 6.3 mins elapsed, rate=71.7k reads per second ||
#> || 90% completed, 6.3 mins elapsed, rate=71.7k reads per second ||
#> || 91% completed, 6.4 mins elapsed, rate=71.6k reads per second ||
#> || 92% completed, 6.5 mins elapsed, rate=71.5k reads per second ||
#> || Start read mapping in chunk. ||
#> || 93% completed, 6.6 mins elapsed, rate=85.3k reads per second ||
#> || 97% completed, 6.8 mins elapsed, rate=71.8k reads per second ||
#> || 98% completed, 6.8 mins elapsed, rate=71.7k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=71.7k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=71.7k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=71.7k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=71.7k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=71.6k reads per second ||
#> || 99% completed, 7.0 mins elapsed, rate=71.6k reads per second ||
#> || 99% completed, 7.0 mins elapsed, rate=71.6k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 30,000,000 ||
#> || Mapped : 5,623,606 (18.7%) ||
#> || Uniquely mapped : 5,623,606 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 24,376,394 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 7.7 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.15M_CC_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.15M_CC_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 19:07:04, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=101.1k reads per second ||
#> || 6% completed, 1.4 mins elapsed, rate=115.2k reads per second ||
#> || 13% completed, 1.7 mins elapsed, rate=115.1k reads per second ||
#> || 16% completed, 1.9 mins elapsed, rate=43.0k reads per second ||
#> || 17% completed, 2.0 mins elapsed, rate=43.6k reads per second ||
#> || 17% completed, 2.0 mins elapsed, rate=44.2k reads per second ||
#> || 18% completed, 2.1 mins elapsed, rate=44.7k reads per second ||
#> || 19% completed, 2.1 mins elapsed, rate=45.2k reads per second ||
#> || 20% completed, 2.2 mins elapsed, rate=45.7k reads per second ||
#> || 20% completed, 2.3 mins elapsed, rate=46.2k reads per second ||
#> || 21% completed, 2.3 mins elapsed, rate=46.6k reads per second ||
#> || 22% completed, 2.4 mins elapsed, rate=47.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 23% completed, 2.6 mins elapsed, rate=83.0k reads per second ||
#> || 29% completed, 2.8 mins elapsed, rate=89.0k reads per second ||
#> || 36% completed, 3.1 mins elapsed, rate=93.3k reads per second ||
#> || 39% completed, 3.3 mins elapsed, rate=60.5k reads per second ||
#> || 40% completed, 3.3 mins elapsed, rate=60.6k reads per second ||
#> || 41% completed, 3.4 mins elapsed, rate=60.6k reads per second ||
#> || 41% completed, 3.5 mins elapsed, rate=60.7k reads per second ||
#> || 42% completed, 3.5 mins elapsed, rate=60.7k reads per second ||
#> || 43% completed, 3.6 mins elapsed, rate=60.7k reads per second ||
#> || 44% completed, 3.6 mins elapsed, rate=60.8k reads per second ||
#> || 45% completed, 3.7 mins elapsed, rate=60.8k reads per second ||
#> || 45% completed, 3.8 mins elapsed, rate=60.8k reads per second ||
#> || Start read mapping in chunk. ||
#> || 46% completed, 3.9 mins elapsed, rate=84.8k reads per second ||
#> || 53% completed, 4.2 mins elapsed, rate=88.0k reads per second ||
#> || 59% completed, 4.5 mins elapsed, rate=90.6k reads per second ||
#> || 62% completed, 4.6 mins elapsed, rate=68.1k reads per second ||
#> || 63% completed, 4.7 mins elapsed, rate=68.1k reads per second ||
#> || 64% completed, 4.7 mins elapsed, rate=68.0k reads per second ||
#> || 65% completed, 4.8 mins elapsed, rate=67.9k reads per second ||
#> || 65% completed, 4.9 mins elapsed, rate=67.9k reads per second ||
#> || 66% completed, 4.9 mins elapsed, rate=67.8k reads per second ||
#> || 67% completed, 5.0 mins elapsed, rate=67.7k reads per second ||
#> || 68% completed, 5.0 mins elapsed, rate=67.7k reads per second ||
#> || 69% completed, 5.1 mins elapsed, rate=67.6k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 5.2 mins elapsed, rate=85.4k reads per second ||
#> || 76% completed, 5.5 mins elapsed, rate=87.6k reads per second ||
#> || 83% completed, 5.8 mins elapsed, rate=89.4k reads per second ||
#> || 86% completed, 6.0 mins elapsed, rate=72.2k reads per second ||
#> || 86% completed, 6.0 mins elapsed, rate=72.1k reads per second ||
#> || 87% completed, 6.1 mins elapsed, rate=72.0k reads per second ||
#> || 88% completed, 6.2 mins elapsed, rate=72.0k reads per second ||
#> || 89% completed, 6.2 mins elapsed, rate=71.9k reads per second ||
#> || 90% completed, 6.3 mins elapsed, rate=71.8k reads per second ||
#> || 90% completed, 6.3 mins elapsed, rate=71.7k reads per second ||
#> || 91% completed, 6.4 mins elapsed, rate=71.6k reads per second ||
#> || 92% completed, 6.5 mins elapsed, rate=71.5k reads per second ||
#> || Start read mapping in chunk. ||
#> || 93% completed, 6.6 mins elapsed, rate=85.7k reads per second ||
#> || 97% completed, 6.8 mins elapsed, rate=72.0k reads per second ||
#> || 98% completed, 6.8 mins elapsed, rate=71.9k reads per second ||
#> || 98% completed, 6.8 mins elapsed, rate=71.9k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=71.9k reads per second ||
#> || 98% completed, 6.9 mins elapsed, rate=71.9k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=71.8k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=71.8k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=71.8k reads per second ||
#> || 99% completed, 7.0 mins elapsed, rate=71.8k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 30,000,000 ||
#> || Mapped : 5,627,855 (18.8%) ||
#> || Uniquely mapped : 5,627,855 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 24,372,145 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 7.7 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.15M_CT_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.15M_CT_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 19:14:45, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=102.2k reads per second ||
#> || 6% completed, 1.4 mins elapsed, rate=114.9k reads per second ||
#> || 13% completed, 1.7 mins elapsed, rate=115.1k reads per second ||
#> || 16% completed, 1.9 mins elapsed, rate=43.3k reads per second ||
#> || 16% completed, 1.9 mins elapsed, rate=43.9k reads per second ||
#> || 17% completed, 2.0 mins elapsed, rate=44.5k reads per second ||
#> || 18% completed, 2.1 mins elapsed, rate=45.0k reads per second ||
#> || 19% completed, 2.1 mins elapsed, rate=45.5k reads per second ||
#> || 20% completed, 2.2 mins elapsed, rate=46.0k reads per second ||
#> || 20% completed, 2.3 mins elapsed, rate=46.4k reads per second ||
#> || 21% completed, 2.3 mins elapsed, rate=46.8k reads per second ||
#> || 22% completed, 2.4 mins elapsed, rate=47.3k reads per second ||
#> || Start read mapping in chunk. ||
#> || 23% completed, 2.5 mins elapsed, rate=83.9k reads per second ||
#> || 29% completed, 2.8 mins elapsed, rate=89.7k reads per second ||
#> || 36% completed, 3.1 mins elapsed, rate=94.0k reads per second ||
#> || 39% completed, 3.2 mins elapsed, rate=60.9k reads per second ||
#> || 40% completed, 3.3 mins elapsed, rate=60.9k reads per second ||
#> || 40% completed, 3.4 mins elapsed, rate=60.9k reads per second ||
#> || 41% completed, 3.4 mins elapsed, rate=61.0k reads per second ||
#> || 42% completed, 3.5 mins elapsed, rate=61.0k reads per second ||
#> || 43% completed, 3.6 mins elapsed, rate=61.1k reads per second ||
#> || 44% completed, 3.6 mins elapsed, rate=61.1k reads per second ||
#> || 44% completed, 3.7 mins elapsed, rate=61.2k reads per second ||
#> || 45% completed, 3.7 mins elapsed, rate=61.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 46% completed, 3.9 mins elapsed, rate=85.5k reads per second ||
#> || 53% completed, 4.2 mins elapsed, rate=88.6k reads per second ||
#> || 59% completed, 4.4 mins elapsed, rate=91.1k reads per second ||
#> || 62% completed, 4.6 mins elapsed, rate=68.5k reads per second ||
#> || 63% completed, 4.6 mins elapsed, rate=68.5k reads per second ||
#> || 64% completed, 4.7 mins elapsed, rate=68.4k reads per second ||
#> || 65% completed, 4.8 mins elapsed, rate=68.3k reads per second ||
#> || 65% completed, 4.8 mins elapsed, rate=68.3k reads per second ||
#> || 66% completed, 4.9 mins elapsed, rate=68.2k reads per second ||
#> || 67% completed, 5.0 mins elapsed, rate=68.2k reads per second ||
#> || 68% completed, 5.0 mins elapsed, rate=68.1k reads per second ||
#> || 68% completed, 5.1 mins elapsed, rate=68.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 5.2 mins elapsed, rate=86.0k reads per second ||
#> || 76% completed, 5.5 mins elapsed, rate=88.1k reads per second ||
#> || 83% completed, 5.8 mins elapsed, rate=90.0k reads per second ||
#> || 86% completed, 5.9 mins elapsed, rate=72.7k reads per second ||
#> || 86% completed, 6.0 mins elapsed, rate=72.6k reads per second ||
#> || 87% completed, 6.0 mins elapsed, rate=72.5k reads per second ||
#> || 88% completed, 6.1 mins elapsed, rate=72.5k reads per second ||
#> || 89% completed, 6.2 mins elapsed, rate=72.4k reads per second ||
#> || 89% completed, 6.2 mins elapsed, rate=72.3k reads per second ||
#> || 90% completed, 6.3 mins elapsed, rate=72.2k reads per second ||
#> || 91% completed, 6.4 mins elapsed, rate=72.1k reads per second ||
#> || 92% completed, 6.4 mins elapsed, rate=72.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 93% completed, 6.5 mins elapsed, rate=86.3k reads per second ||
#> || 97% completed, 6.8 mins elapsed, rate=72.5k reads per second ||
#> || 98% completed, 6.8 mins elapsed, rate=72.5k reads per second ||
#> || 98% completed, 6.8 mins elapsed, rate=72.5k reads per second ||
#> || 98% completed, 6.8 mins elapsed, rate=72.4k reads per second ||
#> || 98% completed, 6.8 mins elapsed, rate=72.4k reads per second ||
#> || 99% completed, 6.8 mins elapsed, rate=72.4k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=72.4k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=72.3k reads per second ||
#> || 99% completed, 6.9 mins elapsed, rate=72.3k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 30,000,000 ||
#> || Mapped : 5,625,823 (18.8%) ||
#> || Uniquely mapped : 5,625,823 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 24,374,177 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 7.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.20M_AA_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.20M_AA_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 19:22:27, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=98.8k reads per second ||
#> || 6% completed, 1.5 mins elapsed, rate=108.9k reads per second ||
#> || 12% completed, 1.9 mins elapsed, rate=42.4k reads per second ||
#> || 12% completed, 2.0 mins elapsed, rate=43.2k reads per second ||
#> || 13% completed, 2.0 mins elapsed, rate=44.0k reads per second ||
#> || 13% completed, 2.1 mins elapsed, rate=44.7k reads per second ||
#> || 14% completed, 2.1 mins elapsed, rate=45.4k reads per second ||
#> || 15% completed, 2.2 mins elapsed, rate=46.1k reads per second ||
#> || 15% completed, 2.2 mins elapsed, rate=46.8k reads per second ||
#> || 16% completed, 2.3 mins elapsed, rate=47.4k reads per second ||
#> || 16% completed, 2.3 mins elapsed, rate=48.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 17% completed, 2.5 mins elapsed, rate=84.6k reads per second ||
#> || 24% completed, 2.9 mins elapsed, rate=90.9k reads per second ||
#> || 29% completed, 3.3 mins elapsed, rate=60.4k reads per second ||
#> || 30% completed, 3.3 mins elapsed, rate=60.7k reads per second ||
#> || 30% completed, 3.4 mins elapsed, rate=60.9k reads per second ||
#> || 31% completed, 3.4 mins elapsed, rate=61.1k reads per second ||
#> || 31% completed, 3.5 mins elapsed, rate=61.3k reads per second ||
#> || 32% completed, 3.5 mins elapsed, rate=61.5k reads per second ||
#> || 33% completed, 3.6 mins elapsed, rate=61.7k reads per second ||
#> || 33% completed, 3.6 mins elapsed, rate=61.9k reads per second ||
#> || 34% completed, 3.7 mins elapsed, rate=62.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 3.9 mins elapsed, rate=86.4k reads per second ||
#> || 41% completed, 4.3 mins elapsed, rate=89.8k reads per second ||
#> || 47% completed, 4.6 mins elapsed, rate=68.4k reads per second ||
#> || 47% completed, 4.6 mins elapsed, rate=68.5k reads per second ||
#> || 48% completed, 4.7 mins elapsed, rate=68.6k reads per second ||
#> || 48% completed, 4.8 mins elapsed, rate=68.7k reads per second ||
#> || 49% completed, 4.8 mins elapsed, rate=68.7k reads per second ||
#> || 49% completed, 4.9 mins elapsed, rate=68.8k reads per second ||
#> || 50% completed, 4.9 mins elapsed, rate=68.9k reads per second ||
#> || 51% completed, 5.0 mins elapsed, rate=68.9k reads per second ||
#> || 51% completed, 5.0 mins elapsed, rate=69.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 52% completed, 5.2 mins elapsed, rate=87.1k reads per second ||
#> || 58% completed, 5.6 mins elapsed, rate=89.3k reads per second ||
#> || 64% completed, 5.9 mins elapsed, rate=72.8k reads per second ||
#> || 65% completed, 6.0 mins elapsed, rate=72.9k reads per second ||
#> || 65% completed, 6.0 mins elapsed, rate=72.9k reads per second ||
#> || 66% completed, 6.1 mins elapsed, rate=72.9k reads per second ||
#> || 66% completed, 6.1 mins elapsed, rate=72.9k reads per second ||
#> || 67% completed, 6.2 mins elapsed, rate=73.0k reads per second ||
#> || 67% completed, 6.2 mins elapsed, rate=73.0k reads per second ||
#> || 68% completed, 6.3 mins elapsed, rate=73.0k reads per second ||
#> || 69% completed, 6.3 mins elapsed, rate=73.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 6.5 mins elapsed, rate=87.4k reads per second ||
#> || 76% completed, 6.9 mins elapsed, rate=89.1k reads per second ||
#> || 81% completed, 7.2 mins elapsed, rate=75.6k reads per second ||
#> || 82% completed, 7.3 mins elapsed, rate=75.6k reads per second ||
#> || 83% completed, 7.3 mins elapsed, rate=75.6k reads per second ||
#> || 83% completed, 7.4 mins elapsed, rate=75.6k reads per second ||
#> || 84% completed, 7.4 mins elapsed, rate=75.6k reads per second ||
#> || 84% completed, 7.5 mins elapsed, rate=75.6k reads per second ||
#> || 85% completed, 7.5 mins elapsed, rate=75.6k reads per second ||
#> || 86% completed, 7.6 mins elapsed, rate=75.6k reads per second ||
#> || 86% completed, 7.6 mins elapsed, rate=75.6k reads per second ||
#> || Start read mapping in chunk. ||
#> || 87% completed, 7.8 mins elapsed, rate=87.6k reads per second ||
#> || 94% completed, 8.2 mins elapsed, rate=89.0k reads per second ||
#> || 96% completed, 8.3 mins elapsed, rate=76.8k reads per second ||
#> || 96% completed, 8.4 mins elapsed, rate=76.8k reads per second ||
#> || 97% completed, 8.4 mins elapsed, rate=76.8k reads per second ||
#> || 97% completed, 8.5 mins elapsed, rate=76.8k reads per second ||
#> || 97% completed, 8.5 mins elapsed, rate=76.8k reads per second ||
#> || 98% completed, 8.5 mins elapsed, rate=76.8k reads per second ||
#> || 98% completed, 8.6 mins elapsed, rate=76.8k reads per second ||
#> || 99% completed, 8.6 mins elapsed, rate=76.8k reads per second ||
#> || 99% completed, 8.6 mins elapsed, rate=76.8k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 39,999,979 ||
#> || Mapped : 28,475,514 (71.2%) ||
#> || Uniquely mapped : 28,475,514 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 11,524,465 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 9.7 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.20M_AC_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.20M_AC_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 19:32:07, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=98.1k reads per second ||
#> || 6% completed, 1.5 mins elapsed, rate=108.8k reads per second ||
#> || 12% completed, 1.9 mins elapsed, rate=42.5k reads per second ||
#> || 12% completed, 2.0 mins elapsed, rate=43.3k reads per second ||
#> || 13% completed, 2.0 mins elapsed, rate=44.1k reads per second ||
#> || 13% completed, 2.1 mins elapsed, rate=44.8k reads per second ||
#> || 14% completed, 2.1 mins elapsed, rate=45.5k reads per second ||
#> || 15% completed, 2.2 mins elapsed, rate=46.2k reads per second ||
#> || 15% completed, 2.2 mins elapsed, rate=46.8k reads per second ||
#> || 16% completed, 2.3 mins elapsed, rate=47.5k reads per second ||
#> || 16% completed, 2.3 mins elapsed, rate=48.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 17% completed, 2.5 mins elapsed, rate=84.7k reads per second ||
#> || 24% completed, 2.9 mins elapsed, rate=91.0k reads per second ||
#> || 29% completed, 3.3 mins elapsed, rate=60.5k reads per second ||
#> || 30% completed, 3.3 mins elapsed, rate=60.8k reads per second ||
#> || 30% completed, 3.4 mins elapsed, rate=61.0k reads per second ||
#> || 31% completed, 3.4 mins elapsed, rate=61.2k reads per second ||
#> || 31% completed, 3.5 mins elapsed, rate=61.4k reads per second ||
#> || 32% completed, 3.5 mins elapsed, rate=61.6k reads per second ||
#> || 33% completed, 3.6 mins elapsed, rate=61.8k reads per second ||
#> || 33% completed, 3.6 mins elapsed, rate=62.0k reads per second ||
#> || 34% completed, 3.7 mins elapsed, rate=62.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 3.9 mins elapsed, rate=85.9k reads per second ||
#> || 41% completed, 4.3 mins elapsed, rate=89.3k reads per second ||
#> || 47% completed, 4.6 mins elapsed, rate=68.2k reads per second ||
#> || 47% completed, 4.7 mins elapsed, rate=68.3k reads per second ||
#> || 48% completed, 4.7 mins elapsed, rate=68.4k reads per second ||
#> || 48% completed, 4.8 mins elapsed, rate=68.5k reads per second ||
#> || 49% completed, 4.8 mins elapsed, rate=68.6k reads per second ||
#> || 50% completed, 4.9 mins elapsed, rate=68.6k reads per second ||
#> || 50% completed, 4.9 mins elapsed, rate=68.7k reads per second ||
#> || 51% completed, 5.0 mins elapsed, rate=68.8k reads per second ||
#> || 51% completed, 5.0 mins elapsed, rate=68.8k reads per second ||
#> || Start read mapping in chunk. ||
#> || 52% completed, 5.2 mins elapsed, rate=86.6k reads per second ||
#> || 59% completed, 5.6 mins elapsed, rate=89.0k reads per second ||
#> || 64% completed, 5.9 mins elapsed, rate=72.7k reads per second ||
#> || 65% completed, 6.0 mins elapsed, rate=72.7k reads per second ||
#> || 65% completed, 6.0 mins elapsed, rate=72.7k reads per second ||
#> || 66% completed, 6.1 mins elapsed, rate=72.7k reads per second ||
#> || 66% completed, 6.1 mins elapsed, rate=72.7k reads per second ||
#> || 67% completed, 6.2 mins elapsed, rate=72.8k reads per second ||
#> || 68% completed, 6.2 mins elapsed, rate=72.8k reads per second ||
#> || 68% completed, 6.3 mins elapsed, rate=72.8k reads per second ||
#> || 69% completed, 6.3 mins elapsed, rate=72.8k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 6.5 mins elapsed, rate=86.7k reads per second ||
#> || 76% completed, 6.9 mins elapsed, rate=88.5k reads per second ||
#> || 82% completed, 7.3 mins elapsed, rate=75.3k reads per second ||
#> || 82% completed, 7.3 mins elapsed, rate=75.3k reads per second ||
#> || 83% completed, 7.4 mins elapsed, rate=75.3k reads per second ||
#> || 83% completed, 7.4 mins elapsed, rate=75.3k reads per second ||
#> || 84% completed, 7.5 mins elapsed, rate=75.3k reads per second ||
#> || 85% completed, 7.5 mins elapsed, rate=75.3k reads per second ||
#> || 85% completed, 7.6 mins elapsed, rate=75.3k reads per second ||
#> || 86% completed, 7.6 mins elapsed, rate=75.3k reads per second ||
#> || 86% completed, 7.7 mins elapsed, rate=75.3k reads per second ||
#> || Start read mapping in chunk. ||
#> || 87% completed, 7.8 mins elapsed, rate=87.1k reads per second ||
#> || 94% completed, 8.2 mins elapsed, rate=88.5k reads per second ||
#> || 96% completed, 8.4 mins elapsed, rate=76.5k reads per second ||
#> || 96% completed, 8.4 mins elapsed, rate=76.5k reads per second ||
#> || 97% completed, 8.5 mins elapsed, rate=76.5k reads per second ||
#> || 97% completed, 8.5 mins elapsed, rate=76.5k reads per second ||
#> || 97% completed, 8.5 mins elapsed, rate=76.5k reads per second ||
#> || 98% completed, 8.6 mins elapsed, rate=76.5k reads per second ||
#> || 98% completed, 8.6 mins elapsed, rate=76.5k reads per second ||
#> || 99% completed, 8.6 mins elapsed, rate=76.5k reads per second ||
#> || 99% completed, 8.7 mins elapsed, rate=76.5k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 39,999,968 ||
#> || Mapped : 28,470,177 (71.2%) ||
#> || Uniquely mapped : 28,470,177 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 11,529,791 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 9.7 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.20M_AT_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.20M_AT_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 19:41:48, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=97.2k reads per second ||
#> || 6% completed, 1.5 mins elapsed, rate=109.1k reads per second ||
#> || 12% completed, 1.9 mins elapsed, rate=42.6k reads per second ||
#> || 12% completed, 2.0 mins elapsed, rate=43.4k reads per second ||
#> || 13% completed, 2.0 mins elapsed, rate=44.2k reads per second ||
#> || 13% completed, 2.1 mins elapsed, rate=45.0k reads per second ||
#> || 14% completed, 2.1 mins elapsed, rate=45.7k reads per second ||
#> || 15% completed, 2.2 mins elapsed, rate=46.3k reads per second ||
#> || 15% completed, 2.2 mins elapsed, rate=47.0k reads per second ||
#> || 16% completed, 2.3 mins elapsed, rate=47.6k reads per second ||
#> || 16% completed, 2.3 mins elapsed, rate=48.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 17% completed, 2.5 mins elapsed, rate=85.8k reads per second ||
#> || 24% completed, 2.9 mins elapsed, rate=91.9k reads per second ||
#> || 29% completed, 3.2 mins elapsed, rate=61.0k reads per second ||
#> || 30% completed, 3.3 mins elapsed, rate=61.2k reads per second ||
#> || 30% completed, 3.4 mins elapsed, rate=61.4k reads per second ||
#> || 31% completed, 3.4 mins elapsed, rate=61.6k reads per second ||
#> || 31% completed, 3.5 mins elapsed, rate=61.8k reads per second ||
#> || 32% completed, 3.5 mins elapsed, rate=62.0k reads per second ||
#> || 33% completed, 3.6 mins elapsed, rate=62.2k reads per second ||
#> || 33% completed, 3.6 mins elapsed, rate=62.4k reads per second ||
#> || 34% completed, 3.7 mins elapsed, rate=62.6k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 3.8 mins elapsed, rate=86.9k reads per second ||
#> || 41% completed, 4.2 mins elapsed, rate=90.3k reads per second ||
#> || 47% completed, 4.6 mins elapsed, rate=68.8k reads per second ||
#> || 47% completed, 4.6 mins elapsed, rate=68.9k reads per second ||
#> || 48% completed, 4.7 mins elapsed, rate=69.0k reads per second ||
#> || 48% completed, 4.7 mins elapsed, rate=69.1k reads per second ||
#> || 49% completed, 4.8 mins elapsed, rate=69.1k reads per second ||
#> || 50% completed, 4.8 mins elapsed, rate=69.2k reads per second ||
#> || 50% completed, 4.9 mins elapsed, rate=69.3k reads per second ||
#> || 51% completed, 4.9 mins elapsed, rate=69.3k reads per second ||
#> || 51% completed, 5.0 mins elapsed, rate=69.4k reads per second ||
#> || Start read mapping in chunk. ||
#> || 52% completed, 5.2 mins elapsed, rate=87.2k reads per second ||
#> || 59% completed, 5.6 mins elapsed, rate=89.4k reads per second ||
#> || 64% completed, 5.9 mins elapsed, rate=73.0k reads per second ||
#> || 65% completed, 6.0 mins elapsed, rate=73.1k reads per second ||
#> || 65% completed, 6.0 mins elapsed, rate=73.1k reads per second ||
#> || 66% completed, 6.1 mins elapsed, rate=73.1k reads per second ||
#> || 66% completed, 6.1 mins elapsed, rate=73.1k reads per second ||
#> || 67% completed, 6.2 mins elapsed, rate=73.1k reads per second ||
#> || 68% completed, 6.2 mins elapsed, rate=73.1k reads per second ||
#> || 68% completed, 6.3 mins elapsed, rate=73.1k reads per second ||
#> || 69% completed, 6.3 mins elapsed, rate=73.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 6.5 mins elapsed, rate=87.4k reads per second ||
#> || 76% completed, 6.9 mins elapsed, rate=89.1k reads per second ||
#> || 82% completed, 7.2 mins elapsed, rate=75.8k reads per second ||
#> || 82% completed, 7.3 mins elapsed, rate=75.8k reads per second ||
#> || 83% completed, 7.3 mins elapsed, rate=75.8k reads per second ||
#> || 83% completed, 7.4 mins elapsed, rate=75.8k reads per second ||
#> || 84% completed, 7.4 mins elapsed, rate=75.8k reads per second ||
#> || 85% completed, 7.5 mins elapsed, rate=75.7k reads per second ||
#> || 85% completed, 7.5 mins elapsed, rate=75.7k reads per second ||
#> || 86% completed, 7.6 mins elapsed, rate=75.7k reads per second ||
#> || 86% completed, 7.6 mins elapsed, rate=75.7k reads per second ||
#> || Start read mapping in chunk. ||
#> || 87% completed, 7.8 mins elapsed, rate=87.4k reads per second ||
#> || 94% completed, 8.2 mins elapsed, rate=88.8k reads per second ||
#> || 96% completed, 8.3 mins elapsed, rate=76.8k reads per second ||
#> || 96% completed, 8.4 mins elapsed, rate=76.8k reads per second ||
#> || 97% completed, 8.4 mins elapsed, rate=76.8k reads per second ||
#> || 97% completed, 8.5 mins elapsed, rate=76.8k reads per second ||
#> || 97% completed, 8.5 mins elapsed, rate=76.8k reads per second ||
#> || 98% completed, 8.5 mins elapsed, rate=76.8k reads per second ||
#> || 98% completed, 8.6 mins elapsed, rate=76.8k reads per second ||
#> || 99% completed, 8.6 mins elapsed, rate=76.8k reads per second ||
#> || 99% completed, 8.6 mins elapsed, rate=76.8k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 39,999,973 ||
#> || Mapped : 28,470,305 (71.2%) ||
#> || Uniquely mapped : 28,470,305 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 11,529,668 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 9.6 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.20M_CA_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.20M_CA_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 19:51:26, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=100.2k reads per second ||
#> || 6% completed, 1.5 mins elapsed, rate=115.0k reads per second ||
#> || 12% completed, 1.9 mins elapsed, rate=43.1k reads per second ||
#> || 12% completed, 2.0 mins elapsed, rate=43.7k reads per second ||
#> || 13% completed, 2.0 mins elapsed, rate=44.3k reads per second ||
#> || 13% completed, 2.1 mins elapsed, rate=44.8k reads per second ||
#> || 14% completed, 2.1 mins elapsed, rate=45.3k reads per second ||
#> || 15% completed, 2.2 mins elapsed, rate=45.8k reads per second ||
#> || 15% completed, 2.3 mins elapsed, rate=46.3k reads per second ||
#> || 16% completed, 2.3 mins elapsed, rate=46.7k reads per second ||
#> || 16% completed, 2.4 mins elapsed, rate=47.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 17% completed, 2.5 mins elapsed, rate=84.4k reads per second ||
#> || 24% completed, 2.9 mins elapsed, rate=91.8k reads per second ||
#> || 29% completed, 3.2 mins elapsed, rate=60.9k reads per second ||
#> || 30% completed, 3.3 mins elapsed, rate=61.0k reads per second ||
#> || 30% completed, 3.4 mins elapsed, rate=61.0k reads per second ||
#> || 31% completed, 3.4 mins elapsed, rate=61.0k reads per second ||
#> || 31% completed, 3.5 mins elapsed, rate=61.1k reads per second ||
#> || 32% completed, 3.6 mins elapsed, rate=61.1k reads per second ||
#> || 33% completed, 3.6 mins elapsed, rate=61.1k reads per second ||
#> || 33% completed, 3.7 mins elapsed, rate=61.2k reads per second ||
#> || 34% completed, 3.7 mins elapsed, rate=61.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 3.9 mins elapsed, rate=85.4k reads per second ||
#> || 41% completed, 4.3 mins elapsed, rate=89.5k reads per second ||
#> || 47% completed, 4.6 mins elapsed, rate=68.5k reads per second ||
#> || 47% completed, 4.7 mins elapsed, rate=68.4k reads per second ||
#> || 48% completed, 4.7 mins elapsed, rate=68.3k reads per second ||
#> || 48% completed, 4.8 mins elapsed, rate=68.3k reads per second ||
#> || 49% completed, 4.8 mins elapsed, rate=68.2k reads per second ||
#> || 50% completed, 4.9 mins elapsed, rate=68.1k reads per second ||
#> || 50% completed, 5.0 mins elapsed, rate=68.1k reads per second ||
#> || 51% completed, 5.0 mins elapsed, rate=68.0k reads per second ||
#> || 51% completed, 5.1 mins elapsed, rate=68.0k reads per second ||
#> || Start read mapping in chunk. ||
#> || 52% completed, 5.2 mins elapsed, rate=85.9k reads per second ||
#> || 59% completed, 5.6 mins elapsed, rate=88.7k reads per second ||
#> || 64% completed, 5.9 mins elapsed, rate=72.7k reads per second ||
#> || 65% completed, 6.0 mins elapsed, rate=72.6k reads per second ||
#> || 65% completed, 6.1 mins elapsed, rate=72.5k reads per second ||
#> || 66% completed, 6.1 mins elapsed, rate=72.4k reads per second ||
#> || 66% completed, 6.2 mins elapsed, rate=72.3k reads per second ||
#> || 67% completed, 6.2 mins elapsed, rate=72.2k reads per second ||
#> || 68% completed, 6.3 mins elapsed, rate=72.1k reads per second ||
#> || 68% completed, 6.4 mins elapsed, rate=72.0k reads per second ||
#> || 69% completed, 6.4 mins elapsed, rate=71.9k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 6.6 mins elapsed, rate=86.1k reads per second ||
#> || 76% completed, 6.9 mins elapsed, rate=88.2k reads per second ||
#> || 82% completed, 7.3 mins elapsed, rate=75.2k reads per second ||
#> || 82% completed, 7.3 mins elapsed, rate=75.1k reads per second ||
#> || 83% completed, 7.4 mins elapsed, rate=75.0k reads per second ||
#> || 83% completed, 7.5 mins elapsed, rate=74.9k reads per second ||
#> || 84% completed, 7.5 mins elapsed, rate=74.8k reads per second ||
#> || 85% completed, 7.6 mins elapsed, rate=74.7k reads per second ||
#> || 85% completed, 7.7 mins elapsed, rate=74.6k reads per second ||
#> || 86% completed, 7.7 mins elapsed, rate=74.5k reads per second ||
#> || 86% completed, 7.8 mins elapsed, rate=74.4k reads per second ||
#> || Start read mapping in chunk. ||
#> || 87% completed, 7.9 mins elapsed, rate=86.1k reads per second ||
#> || 94% completed, 8.3 mins elapsed, rate=87.8k reads per second ||
#> || 96% completed, 8.4 mins elapsed, rate=76.0k reads per second ||
#> || 96% completed, 8.5 mins elapsed, rate=75.9k reads per second ||
#> || 97% completed, 8.5 mins elapsed, rate=75.9k reads per second ||
#> || 97% completed, 8.6 mins elapsed, rate=75.8k reads per second ||
#> || 97% completed, 8.6 mins elapsed, rate=75.7k reads per second ||
#> || 98% completed, 8.7 mins elapsed, rate=75.7k reads per second ||
#> || 98% completed, 8.7 mins elapsed, rate=75.6k reads per second ||
#> || 99% completed, 8.7 mins elapsed, rate=75.5k reads per second ||
#> || 99% completed, 8.8 mins elapsed, rate=75.5k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 39,999,987 ||
#> || Mapped : 7,495,804 (18.7%) ||
#> || Uniquely mapped : 7,495,804 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 32,504,183 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 9.8 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.20M_CC_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.20M_CC_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 20:01:15, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.1 mins elapsed, rate=99.1k reads per second ||
#> || 6% completed, 1.5 mins elapsed, rate=115.2k reads per second ||
#> || 12% completed, 1.9 mins elapsed, rate=43.2k reads per second ||
#> || 12% completed, 1.9 mins elapsed, rate=43.8k reads per second ||
#> || 13% completed, 2.0 mins elapsed, rate=44.4k reads per second ||
#> || 13% completed, 2.1 mins elapsed, rate=44.9k reads per second ||
#> || 14% completed, 2.1 mins elapsed, rate=45.4k reads per second ||
#> || 15% completed, 2.2 mins elapsed, rate=45.9k reads per second ||
#> || 15% completed, 2.3 mins elapsed, rate=46.3k reads per second ||
#> || 16% completed, 2.3 mins elapsed, rate=46.8k reads per second ||
#> || 16% completed, 2.4 mins elapsed, rate=47.2k reads per second ||
#> || Start read mapping in chunk. ||
#> || 17% completed, 2.5 mins elapsed, rate=84.0k reads per second ||
#> || 24% completed, 2.9 mins elapsed, rate=91.5k reads per second ||
#> || 29% completed, 3.2 mins elapsed, rate=61.0k reads per second ||
#> || 30% completed, 3.3 mins elapsed, rate=61.0k reads per second ||
#> || 30% completed, 3.4 mins elapsed, rate=61.0k reads per second ||
#> || 31% completed, 3.4 mins elapsed, rate=61.1k reads per second ||
#> || 31% completed, 3.5 mins elapsed, rate=61.1k reads per second ||
#> || 32% completed, 3.6 mins elapsed, rate=61.2k reads per second ||
#> || 33% completed, 3.6 mins elapsed, rate=61.2k reads per second ||
#> || 33% completed, 3.7 mins elapsed, rate=61.3k reads per second ||
#> || 34% completed, 3.7 mins elapsed, rate=61.3k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 3.9 mins elapsed, rate=85.2k reads per second ||
#> || 41% completed, 4.3 mins elapsed, rate=89.3k reads per second ||
#> || 47% completed, 4.6 mins elapsed, rate=68.4k reads per second ||
#> || 47% completed, 4.7 mins elapsed, rate=68.4k reads per second ||
#> || 48% completed, 4.7 mins elapsed, rate=68.3k reads per second ||
#> || 48% completed, 4.8 mins elapsed, rate=68.2k reads per second ||
#> || 49% completed, 4.8 mins elapsed, rate=68.1k reads per second ||
#> || 49% completed, 4.9 mins elapsed, rate=68.1k reads per second ||
#> || 50% completed, 5.0 mins elapsed, rate=68.0k reads per second ||
#> || 51% completed, 5.0 mins elapsed, rate=68.0k reads per second ||
#> || 51% completed, 5.1 mins elapsed, rate=67.9k reads per second ||
#> || Start read mapping in chunk. ||
#> || 52% completed, 5.2 mins elapsed, rate=85.6k reads per second ||
#> || 58% completed, 5.6 mins elapsed, rate=88.4k reads per second ||
#> || 64% completed, 6.0 mins elapsed, rate=72.4k reads per second ||
#> || 65% completed, 6.0 mins elapsed, rate=72.3k reads per second ||
#> || 65% completed, 6.1 mins elapsed, rate=72.2k reads per second ||
#> || 66% completed, 6.1 mins elapsed, rate=72.2k reads per second ||
#> || 66% completed, 6.2 mins elapsed, rate=72.1k reads per second ||
#> || 67% completed, 6.3 mins elapsed, rate=72.0k reads per second ||
#> || 67% completed, 6.3 mins elapsed, rate=71.9k reads per second ||
#> || 68% completed, 6.4 mins elapsed, rate=71.8k reads per second ||
#> || 69% completed, 6.4 mins elapsed, rate=71.7k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 6.6 mins elapsed, rate=85.7k reads per second ||
#> || 76% completed, 7.0 mins elapsed, rate=87.7k reads per second ||
#> || 81% completed, 7.3 mins elapsed, rate=74.9k reads per second ||
#> || 82% completed, 7.4 mins elapsed, rate=74.8k reads per second ||
#> || 83% completed, 7.4 mins elapsed, rate=74.7k reads per second ||
#> || 83% completed, 7.5 mins elapsed, rate=74.6k reads per second ||
#> || 84% completed, 7.6 mins elapsed, rate=74.5k reads per second ||
#> || 84% completed, 7.6 mins elapsed, rate=74.4k reads per second ||
#> || 85% completed, 7.7 mins elapsed, rate=74.3k reads per second ||
#> || 86% completed, 7.7 mins elapsed, rate=74.2k reads per second ||
#> || 86% completed, 7.8 mins elapsed, rate=74.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 87% completed, 7.9 mins elapsed, rate=85.7k reads per second ||
#> || 94% completed, 8.3 mins elapsed, rate=87.4k reads per second ||
#> || 96% completed, 8.5 mins elapsed, rate=75.8k reads per second ||
#> || 96% completed, 8.5 mins elapsed, rate=75.7k reads per second ||
#> || 97% completed, 8.6 mins elapsed, rate=75.6k reads per second ||
#> || 97% completed, 8.6 mins elapsed, rate=75.6k reads per second ||
#> || 97% completed, 8.6 mins elapsed, rate=75.5k reads per second ||
#> || 98% completed, 8.7 mins elapsed, rate=75.5k reads per second ||
#> || 98% completed, 8.7 mins elapsed, rate=75.4k reads per second ||
#> || 99% completed, 8.8 mins elapsed, rate=75.3k reads per second ||
#> || 99% completed, 8.8 mins elapsed, rate=75.3k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 39,999,981 ||
#> || Mapped : 7,502,630 (18.8%) ||
#> || Uniquely mapped : 7,502,630 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 32,497,351 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 9.8 minutes ||
#> || ||
#> \\============================================================================//
#>
#> [1] "Index directory already exists remove it if you want to recreate it"
#>
#> ========== _____ _ _ ____ _____ ______ _____
#> ===== / ____| | | | _ \| __ \| ____| /\ | __ \
#> ===== | (___ | | | | |_) | |__) | |__ / \ | | | |
#> ==== \___ \| | | | _ <| _ /| __| / /\ \ | | | |
#> ==== ____) | |__| | |_) | | \ \| |____ / ____ \| |__| |
#> ========== |_____/ \____/|____/|_| \_\______/_/ \_\_____/
#> Rsubread 2.20.0
#>
#> //================================= setting ==================================\\
#> || ||
#> || Function : Read alignment (DNA-Seq) ||
#> || Input file : prepro.PPP-BG.20M_CT_valid.fastq.gz ||
#> || Output file : prepro.PPP-BG.20M_CT_valid.fastq.gz.bam (BAM), Sorted ||
#> || Index name : index ||
#> || ||
#> || ------------------------------------ ||
#> || ||
#> || Threads : 1 ||
#> || Phred offset : 33 ||
#> || Min votes : 3 / 10 ||
#> || Max mismatches : 1 ||
#> || Max indel length : 0 ||
#> || Report multi-mapping reads : no ||
#> || Max alignments per multi-mapping read : 1 ||
#> || ||
#> \\============================================================================//
#>
#> //================ Running (01-Nov-2025 20:11:06, pid=392792) ================\\
#> || ||
#> || Check the input reads. ||
#> || The input file contains base space reads. ||
#> || Initialise the memory objects. ||
#> || Estimate the mean read length. ||
#> || WARNING - The specified Phred score offset (33) seems incorrect. ||
#> || ASCII values of the quality scores of read bases included in ||
#> || the first 3000 reads were found to be within the range of ||
#> || [63,63]. ||
#> || ||
#> || Create the output BAM file. ||
#> || Check the index. ||
#> || Init the voting space. ||
#> || Global environment is initialised. ||
#> || Load the 1-th index block... ||
#> || The index block has been loaded. ||
#> || Start read mapping in chunk. ||
#> || 0% completed, 1.2 mins elapsed, rate=100.3k reads per second ||
#> || 6% completed, 1.5 mins elapsed, rate=116.0k reads per second ||
#> || 12% completed, 1.9 mins elapsed, rate=43.2k reads per second ||
#> || 12% completed, 2.0 mins elapsed, rate=43.8k reads per second ||
#> || 13% completed, 2.0 mins elapsed, rate=44.4k reads per second ||
#> || 13% completed, 2.1 mins elapsed, rate=44.9k reads per second ||
#> || 14% completed, 2.1 mins elapsed, rate=45.4k reads per second ||
#> || 15% completed, 2.2 mins elapsed, rate=45.8k reads per second ||
#> || 15% completed, 2.3 mins elapsed, rate=46.3k reads per second ||
#> || 16% completed, 2.3 mins elapsed, rate=46.7k reads per second ||
#> || 16% completed, 2.4 mins elapsed, rate=47.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 17% completed, 2.5 mins elapsed, rate=83.7k reads per second ||
#> || 24% completed, 2.9 mins elapsed, rate=91.1k reads per second ||
#> || 29% completed, 3.3 mins elapsed, rate=60.8k reads per second ||
#> || 30% completed, 3.3 mins elapsed, rate=60.8k reads per second ||
#> || 30% completed, 3.4 mins elapsed, rate=60.9k reads per second ||
#> || 31% completed, 3.4 mins elapsed, rate=60.9k reads per second ||
#> || 31% completed, 3.5 mins elapsed, rate=61.0k reads per second ||
#> || 32% completed, 3.6 mins elapsed, rate=61.0k reads per second ||
#> || 33% completed, 3.6 mins elapsed, rate=61.1k reads per second ||
#> || 33% completed, 3.7 mins elapsed, rate=61.1k reads per second ||
#> || 34% completed, 3.7 mins elapsed, rate=61.1k reads per second ||
#> || Start read mapping in chunk. ||
#> || 34% completed, 3.9 mins elapsed, rate=85.3k reads per second ||
#> || 41% completed, 4.3 mins elapsed, rate=89.4k reads per second ||
#> || 47% completed, 4.6 mins elapsed, rate=68.4k reads per second ||
#> || 47% completed, 4.7 mins elapsed, rate=68.4k reads per second ||
#> || 48% completed, 4.7 mins elapsed, rate=68.3k reads per second ||
#> || 48% completed, 4.8 mins elapsed, rate=68.2k reads per second ||
#> || 49% completed, 4.8 mins elapsed, rate=68.2k reads per second ||
#> || 49% completed, 4.9 mins elapsed, rate=68.1k reads per second ||
#> || 50% completed, 5.0 mins elapsed, rate=68.1k reads per second ||
#> || 51% completed, 5.0 mins elapsed, rate=68.0k reads per second ||
#> || 51% completed, 5.1 mins elapsed, rate=67.9k reads per second ||
#> || Start read mapping in chunk. ||
#> || 52% completed, 5.2 mins elapsed, rate=85.7k reads per second ||
#> || 58% completed, 5.6 mins elapsed, rate=88.5k reads per second ||
#> || 64% completed, 5.9 mins elapsed, rate=72.5k reads per second ||
#> || 65% completed, 6.0 mins elapsed, rate=72.4k reads per second ||
#> || 65% completed, 6.1 mins elapsed, rate=72.3k reads per second ||
#> || 66% completed, 6.1 mins elapsed, rate=72.2k reads per second ||
#> || 66% completed, 6.2 mins elapsed, rate=72.2k reads per second ||
#> || 67% completed, 6.2 mins elapsed, rate=72.1k reads per second ||
#> || 67% completed, 6.3 mins elapsed, rate=72.0k reads per second ||
#> || 68% completed, 6.4 mins elapsed, rate=71.9k reads per second ||
#> || 69% completed, 6.4 mins elapsed, rate=71.9k reads per second ||
#> || Start read mapping in chunk. ||
#> || 69% completed, 6.6 mins elapsed, rate=86.0k reads per second ||
#> || 76% completed, 6.9 mins elapsed, rate=88.1k reads per second ||
#> || 82% completed, 7.3 mins elapsed, rate=75.1k reads per second ||
#> || 82% completed, 7.3 mins elapsed, rate=75.0k reads per second ||
#> || 83% completed, 7.4 mins elapsed, rate=74.9k reads per second ||
#> || 83% completed, 7.5 mins elapsed, rate=74.8k reads per second ||
#> || 84% completed, 7.5 mins elapsed, rate=74.7k reads per second ||
#> || 84% completed, 7.6 mins elapsed, rate=74.6k reads per second ||
#> || 85% completed, 7.7 mins elapsed, rate=74.5k reads per second ||
#> || 86% completed, 7.7 mins elapsed, rate=74.4k reads per second ||
#> || 86% completed, 7.8 mins elapsed, rate=74.4k reads per second ||
#> || Start read mapping in chunk. ||
#> || 87% completed, 7.9 mins elapsed, rate=86.0k reads per second ||
#> || 94% completed, 8.3 mins elapsed, rate=87.8k reads per second ||
#> || 96% completed, 8.4 mins elapsed, rate=76.0k reads per second ||
#> || 96% completed, 8.5 mins elapsed, rate=75.9k reads per second ||
#> || 97% completed, 8.5 mins elapsed, rate=75.9k reads per second ||
#> || 97% completed, 8.6 mins elapsed, rate=75.8k reads per second ||
#> || 97% completed, 8.6 mins elapsed, rate=75.7k reads per second ||
#> || 98% completed, 8.7 mins elapsed, rate=75.7k reads per second ||
#> || 98% completed, 8.7 mins elapsed, rate=75.6k reads per second ||
#> || 99% completed, 8.8 mins elapsed, rate=75.6k reads per second ||
#> || 99% completed, 8.8 mins elapsed, rate=75.5k reads per second ||
#> || ||
#> || Completed successfully. ||
#> || ||
#> \\==================================== ====================================//
#>
#> //================================ Summary =================================\\
#> || ||
#> || Total reads : 39,999,991 ||
#> || Mapped : 7,503,456 (18.8%) ||
#> || Uniquely mapped : 7,503,456 ||
#> || Multi-mapping : 0 ||
#> || ||
#> || Unmapped : 32,496,535 ||
#> || ||
#> || Indels : 0 ||
#> || ||
#> || WARNING : Phred offset (33) incorrect? ||
#> || ||
#> || Running time : 9.8 minutes ||
#> || ||
#> \\============================================================================//
prw_sr <- prw_sr %>%
t %>%
as_tibble() %>%
select(time=Elapsed_Time_sec,
peak_ram=Peak_RAM_Used_MiB) %>%
mutate(time=duration(as.numeric(time), units="seconds"),
peak_ram = as.numeric(peak_ram),
nb_reads = c(12, 60, 120, 180, 240),
aligner = "subread")
prw_sr %>%
knitr::kable()
time |
peak_ram |
nb_reads |
features |
|---|---|---|---|
573.282000000007s (~9.55 minutes) |
0.6 |
12 |
subread |
1257.19799999999s (~20.95 minutes) |
0.4 |
60 |
subread |
1982.12s (~33.04 minutes) |
0.4 |
120 |
subread |
2751.33399999999s (~45.86 minutes) |
0.4 |
180 |
subread |
3507.849s (~58.46 minutes) |
0.4 |
240 |
subread |
Count table computation: quantify_from_bam_files
Measure time and maximum RAM in use for the quantify_from_bam_files function:
samples <- c("PPP.r1", "PPP.r2", "PPP.r3", "BG.r1", "BG.r2", "BG.r3")
prw_counts <- sapply(1:length(dirout), function(i) {
bam_files <- list.files(
paste0(
list.files(
dirout[i],
pattern = "_valid.fastq.gz$",
full.names = TRUE
),
"_MW2.fasta_mm1_bowtie"
),
pattern=".bam$",
full.names = TRUE
)
peakRAM(
count_table <- quantify_from_bam_files(
bam_files = bam_files,
sample_ids = samples,
fun = quantify_5prime,
force = TRUE
))
}
)
prw_counts <- prw_counts %>%
t %>%
as_tibble() %>%
select(time=Elapsed_Time_sec,
peak_ram=Peak_RAM_Used_MiB) %>%
mutate(time=duration(as.numeric(time), units="seconds"),
peak_ram = as.numeric(peak_ram),
nb_reads = c(12, 60, 120, 180, 240),
quantif = "quantify_5prime")
prw_counts %>%
knitr::kable()
time |
peak_ram |
nb_reads |
features |
|---|---|---|---|
71.3050000000076s (~1.19 minutes) |
476 |
12 |
quantify_5prime |
429.542000000001s (~7.16 minutes) |
1916 |
60 |
quantify_5prime |
947.425999999992s (~15.79 minutes) |
4174 |
120 |
quantify_5prime |
1570.59999999999s (~26.18 minutes) |
6017 |
180 |
quantify_5prime |
2433.204s (~40.55 minutes) |
7977 |
240 |
quantify_5prime |
Mapping and quantification summary
prw_map <- bind_rows(prw_bt, prw_sr, prw_counts)
Running time and memory footprint depending on the number of reads:
p1 <- prw_map %>%
ggplot() +
geom_line(aes(x=nb_reads, y=time, color=features),
show.legend = FALSE) +
geom_point(aes(x=nb_reads, y=time, shape=features, color=features),
show.legend = FALSE) +
theme_light() +
xlab("Number of reads (in millions)") +
ylab("Duration (in seconds)") +
ggtitle("Running times for mapping and quantification")
p2 <- prw_map %>%
ggplot() +
geom_line(aes(x=nb_reads, y=peak_ram, color=features)) +
geom_point(aes(x=nb_reads, y=peak_ram, shape=features, color=features)) +
theme_light() +
xlab("Number of reads (in millions)") +
ylab("Memory used (in MB)") +
ggtitle("Memory used during mapping and quantification") +
theme(legend.position="inside", legend.position.inside = c(0.65, 0.2))
ggarrange(p1, p2)
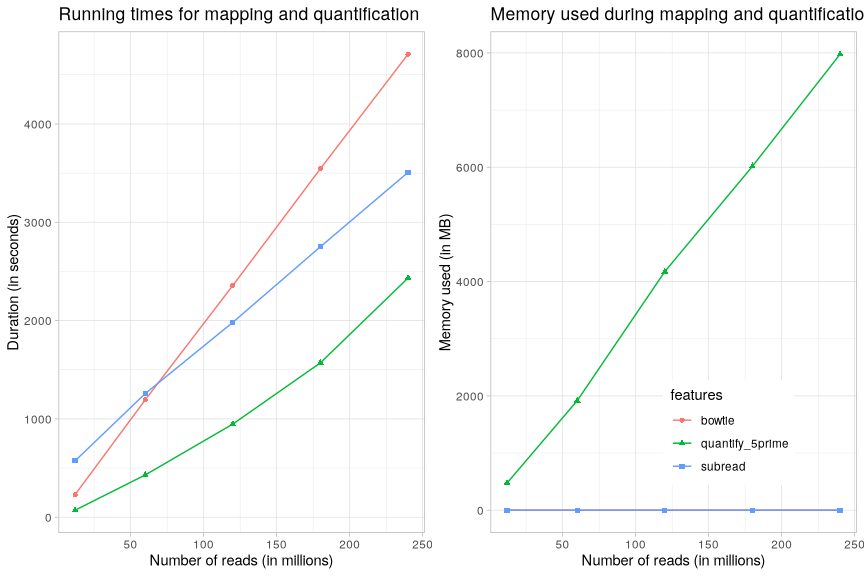
Note
In the above graphs, we can observe that the running times scale linearly with the number of sequencing reads to process.
Regarding the memory footprint, both aligners require a small amount of memory. The quantification to obtain the count table with the quantify_5prime function from rnaends requires an amount of memory that scales linearly with the number of reads as it has to process all reads to compute the 5’ end occurences.
TSS identification: TSS
Load quantifications by re-calling the last quantify_5prime with force=FALSE which will return the previous results.
i <- 5
bam_files <- list.files(
paste0(
list.files(
dirout[i],
pattern = "_valid.fastq.gz$",
full.names = TRUE
),
"_MW2.fasta_mm1_bowtie"
),
pattern=".bam$",
full.names = TRUE
)
count_table <- quantify_from_bam_files(
bam_files = bam_files,
sample_ids = samples,
fun = quantify_5prime,
force = FALSE
)
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_AA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_AA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_AC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_AC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_AT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_AT_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_CA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_CA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_CC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_CC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_CT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_CT_valid.fastq.gz.bam.csv
Design table
design_tb <- tibble(
sample_id = samples,
count_of = c("total", "total", "total", "fraction", "fraction", "fraction"),
replicate = c(1,2,3, 1,2,3)
)
design_tb %>%
knitr::kable()
sample_id |
count_of |
replicate |
|---|---|---|
PPP.r1 |
total |
1 |
PPP.r2 |
total |
2 |
PPP.r3 |
total |
3 |
BG.r1 |
fraction |
1 |
BG.r2 |
fraction |
2 |
BG.r3 |
fraction |
3 |
Measure time and maximum RAM in use for the TSS function:
prw_tss <- sapply(1:length(dirout), function(i) {
bam_files <- list.files(
paste0(
list.files(
dirout[i],
pattern = "_valid.fastq.gz$",
full.names = TRUE
),
"_MW2.fasta_mm1_bowtie"
),
pattern=".bam$",
full.names = TRUE
)
peakRAM(
TSS(
quantify_from_bam_files(
bam_files = bam_files,
sample_ids = samples,
fun = quantify_5prime,
force = FALSE
),
design_tb))
}
)
#> Quantification 5' output file exists, set force=T to overwrite: quantif.1M/prepro.PPP-BG.1M_AA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.1M_AA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.1M/prepro.PPP-BG.1M_AC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.1M_AC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.1M/prepro.PPP-BG.1M_AT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.1M_AT_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.1M/prepro.PPP-BG.1M_CA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.1M_CA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.1M/prepro.PPP-BG.1M_CC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.1M_CC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.1M/prepro.PPP-BG.1M_CT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.1M_CT_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.5M/prepro.PPP-BG.5M_AA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.5M_AA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.5M/prepro.PPP-BG.5M_AC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.5M_AC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.5M/prepro.PPP-BG.5M_AT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.5M_AT_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.5M/prepro.PPP-BG.5M_CA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.5M_CA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.5M/prepro.PPP-BG.5M_CC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.5M_CC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.5M/prepro.PPP-BG.5M_CT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.5M_CT_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.10M/prepro.PPP-BG.10M_AA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.10M_AA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.10M/prepro.PPP-BG.10M_AC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.10M_AC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.10M/prepro.PPP-BG.10M_AT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.10M_AT_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.10M/prepro.PPP-BG.10M_CA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.10M_CA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.10M/prepro.PPP-BG.10M_CC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.10M_CC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.10M/prepro.PPP-BG.10M_CT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.10M_CT_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.15M/prepro.PPP-BG.15M_AA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.15M_AA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.15M/prepro.PPP-BG.15M_AC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.15M_AC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.15M/prepro.PPP-BG.15M_AT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.15M_AT_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.15M/prepro.PPP-BG.15M_CA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.15M_CA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.15M/prepro.PPP-BG.15M_CC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.15M_CC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.15M/prepro.PPP-BG.15M_CT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.15M_CT_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_AA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_AA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_AC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_AC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_AT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_AT_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_CA_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_CA_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_CC_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_CC_valid.fastq.gz.bam.csv
#> Quantification 5' output file exists, set force=T to overwrite: quantif.20M/prepro.PPP-BG.20M_CT_valid.fastq.gz_MW2.fasta_mm1_bowtie/prepro.PPP-BG.20M_CT_valid.fastq.gz.bam.csv
prw_tss <- prw_tss %>%
t %>%
as_tibble() %>%
select(time=Elapsed_Time_sec,
peak_ram=Peak_RAM_Used_MiB) %>%
mutate(time=duration(as.numeric(time), units="seconds"),
peak_ram = as.numeric(peak_ram),
nb_reads = c(12, 60, 120, 180, 240),
analysis = "TSS")
prw_tss %>%
knitr::kable()
time |
peak_ram |
nb_reads |
features |
|---|---|---|---|
4.18499999999767s |
2206 |
12 |
TSS |
25.1370000000024s |
1263 |
60 |
TSS |
52.4130000000005s |
924 |
120 |
TSS |
75.1419999999925s (~1.25 minutes) |
924 |
180 |
TSS |
93.3399999999965s (~1.56 minutes) |
924 |
240 |
TSS |
Summary
p1 <- prw_tss %>%
ggplot() +
geom_line(aes(x=nb_reads, y=time, color=features),
show.legend = FALSE) +
geom_point(aes(x=nb_reads, y=time, shape=features, color=features),
show.legend = FALSE) +
theme_light() +
xlab("Number of reads (in millions)") +
ylab("Duration (in seconds)") +
ggtitle("Running times for TSS identification")
p2 <- prw_tss %>%
ggplot() +
geom_line(aes(x=nb_reads, y=peak_ram, color=features),
show.legend = FALSE) +
geom_point(aes(x=nb_reads, y=peak_ram, shape=features, color=features),
show.legend = FALSE) +
theme_light() +
xlab("Number of reads (in millions)") +
ylab("Memory used (in MB)") +
ggtitle("Memory used during TSS identification")
ggarrange(p1, p2)
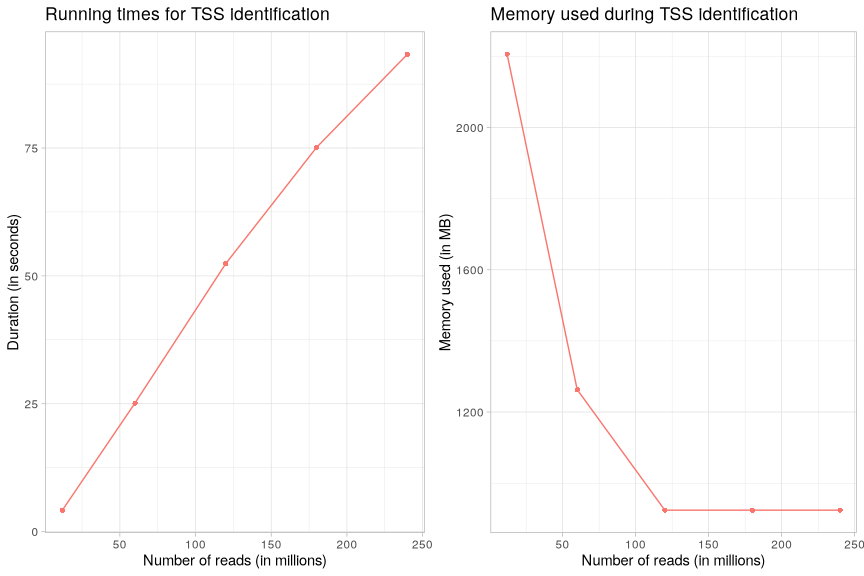
Note
In the above graphs, we can observe that the running times scale linearly with the number of reads processed. Regarding the memory footprint, once the count table is computed in the previous analysis step, we can observe that the tibble representation used its storage is around 1-2GB.
mem_used()
#> 1.15 GB